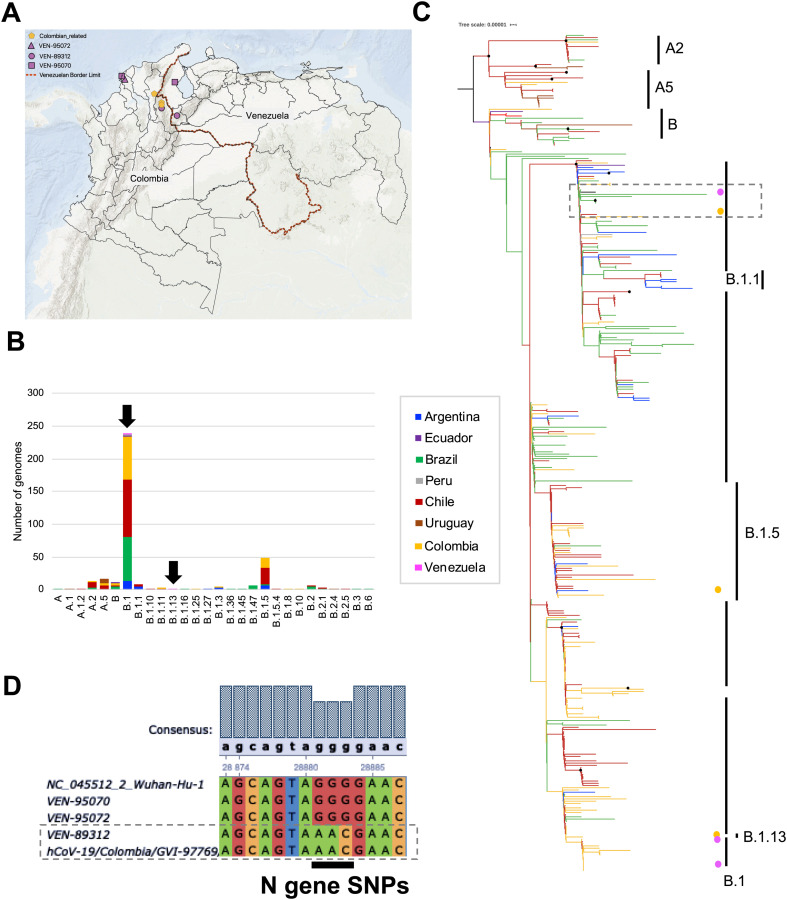
Fig. 1.
Regional comparative genomic analysis of SARS-CoV-2. A. Geographical distribution of the sequences from Venezuelan genomes and the available Colombian genomes analyzed in this study. The pink symbols indicate the Venezuelan patients identified in Colombia with their respective Venezuelan state origin. B. Stacked bar plot of the number of genomes per lineage determined using Phylogenetic Assignment of Named Global Outbreak LINeages ‘Pangolin’ tool. Three SARS-CoV-2 genomes from Venezuela were compared with 376 assemblies from other 7 South American countries (Argentina, Brazil, Chile, Colombia, Ecuador, Peru and Uruguay) using the publicly available GISAID EpiCoV™ database (https://www.gisaid.org/). Frequencies are discriminated by country of origin. The Venezuelan lineages are identified by black arrows. C. Maximum likelihood tree built in IQtree shows the phylogenetic relationships between genomes from Venezuela (pink dots) and the closest Colombian regions (yellow dots) with other South American genomes. The branches were colored according to the country of origin, using the color code of A panel. The clustering of the most frequent pangolin lineages (n > 10) is represented on the right side. The black dots represent highly supported nodes. D. Multiple alignment of the Nucleocapsid gene showing the substitutions found using the Wuhan-1 sequence as reference.