Fig 1.
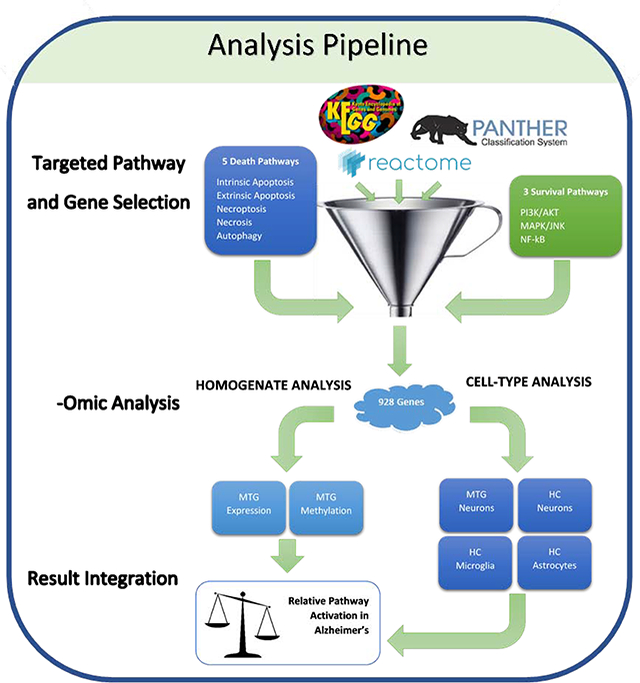
Hypothesis-Testing Analysis Pipeline. Curated genes from three databases (KEGG, Reactome, and PANTHER) were retrieved for eight pathways selected for their role in either promoting or hindering cell death. These unique 928 genes of interest were then analyzed in one of six datasets that can be divided into two wings: 1) analysis of homogenate expression and methylation data from the middle temporal gyrus (MTG) and, 2), cell type specific expression data from LCM cells from the MTG and hippocampus (HC). From these six datasets, the methylation/expression levels from only the significant genes of interest were compared between the datasets. Relative pathway activation in AD was inferred by the methylation/expression status of significant components contributed by each of the six datasets, allowing for a pathway-rather than gene-centric interpretation of the results.
