Fig. 4.
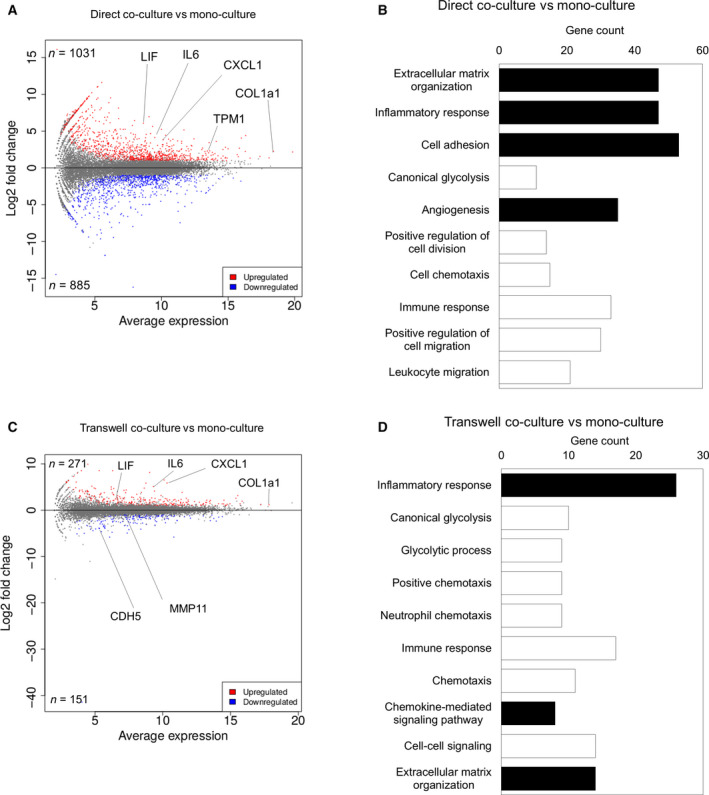
Global gene expression pattern showed that AD‐MSCs were differentiated into CAF‐like cells in vitro. (A) MA plot, a scatter plot of log2 fold change versus the average expression, showing differentially expressed genes (adjusted P‐value < 0.05 and log2 [fold change] ≥ 1) in a direct co‐cultured AD‐MSCs compared to those in monocultured AD‐MSCs. Upregulated genes are shown in red, and downregulated genes are shown in blue. (B) GO enrichment analysis of differentially expressed genes in direct co‐cultured AD‐MSCs compared to those in monocultured AD‐MSC. CAF‐related GO terms are shown in black. (C) MA plot, a scatter plot of log2 fold change versus the average expression, showing differentially expressed genes (adjusted P‐value < 0.05 and log2 [fold change] ≥ 1) in transwell co‐cultured AD‐MSCs compared to those in monocultured AD‐MSCs. Upregulated genes are shown in red, and downregulated genes are shown in blue. (D) GO enrichment analysis of differentially expressed genes in transwell co‐cultured AD‐MSCs compared to those in monocultured AD‐MSC. CAF‐related GO terms are shown in black.
