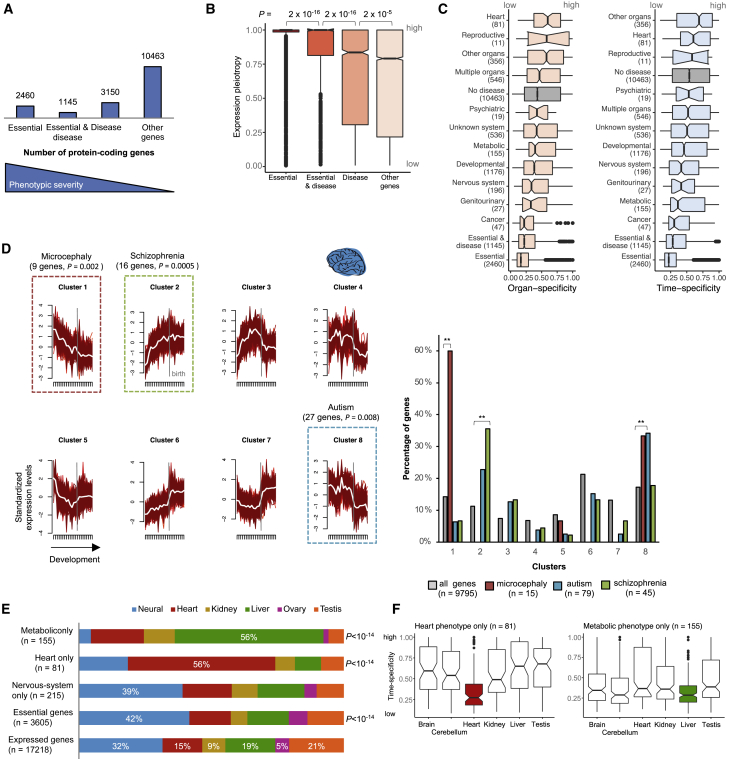
Figure 2.
Spatiotemporal Profiles of Disease Genes
(A) Number of expressed (RPKM >1) protein-coding genes in different classes of phenotypic severity.
(B) Expression pleiotropy of genes in different classes of phenotypic severity (p values from Wilcoxon rank sum test, two sided; the number of genes in each class is shown in A).
(C) Organ and time specificity (median across organs) of genes associated with different disease classes (number of genes in each class in parentheses). Genes are only assigned to one class; those affecting multiple organs appear in the classes “Multiple organs,” “Developmental”, or “Other organs” depending on whether they affect multiple organs that include at least one of the organs in this study, are associated with developmental phenotypes, or affect organs that are not part of this study, respectively.
(D) Genes associated with primary microcephaly, autism, and schizophrenia are significantly enriched in distinct expression clusters in the brain (binomial tests with Bonferroni correction). On the left are the developmental profiles for the clusters identified through soft clustering of the brain developmental samples. The y axis shows standardized expression levels (STAR Methods). The profile of each gene in the cluster is shown in red; the white line shows the cluster center. The genes associated with each disorder are significantly enriched in only one of the eight clusters (right).
(E) Organs where genes associated with organ-specific phenotypes show maximum expression. p values are from binomial tests after Bonferroni correction.
(F) Time specificity in the different organs of genes with heart- and metabolic-specific phenotypes.
In (B), (C), and (F), the boxplots depict the median ±25th and 75th percentiles, with the whiskers at 1.5 times the interquartile range.