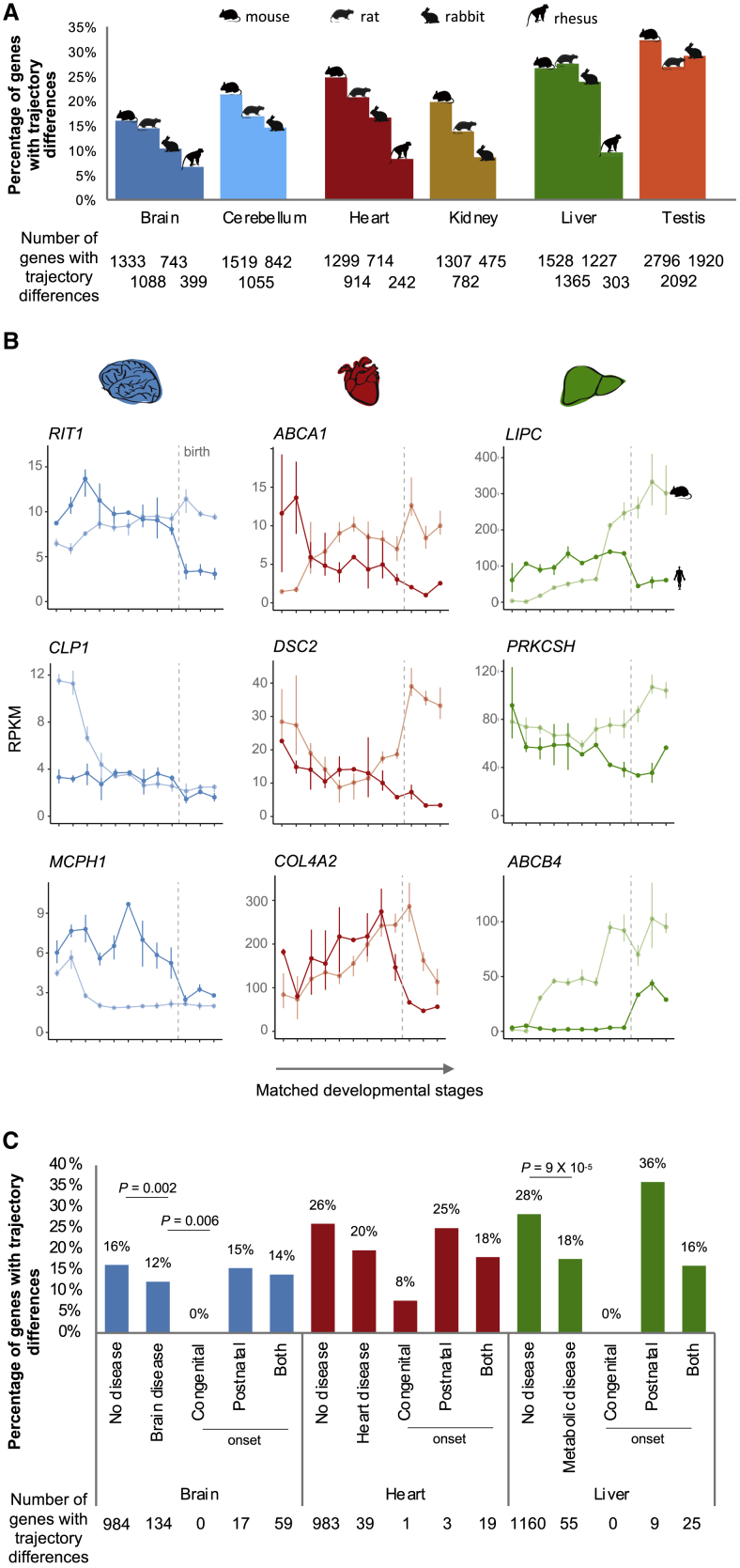
Figure 6.
Developmental Trajectory Differences
(A) Percentage of genes in each organ that have different trajectories between human and mouse, rat, rabbit, and rhesus macaque (because of the shorter time series, this analysis was only performed for brain, heart, and liver in rhesus). The sets of genes compared differ between species as they correspond to the total number of 1:1 orthologs available between human and each species. Figure S6C shows the same analysis but using the same set of 1:1 orthologs across all species (similar results). Table S3 lists the genes tested for trajectory differences and those found to be significantly different between human and each of the other species.
(B) Examples of human disease genes with different developmental trajectories between human and mouse in the affected organ (FDR <5%).
(C) Percentage of genes in brain, heart, and liver that differ in trajectories between human and mouse. Bonferroni-corrected p values for comparisons between disease and non-disease genes are from Fisher’s exact tests, and Bonferroni-corrected p values for comparisons of disease genes with different ages of onset are from binomial tests.