Figure 1.
Termination of snRNA Transcription Is Partially Impaired by Integrator Depletion
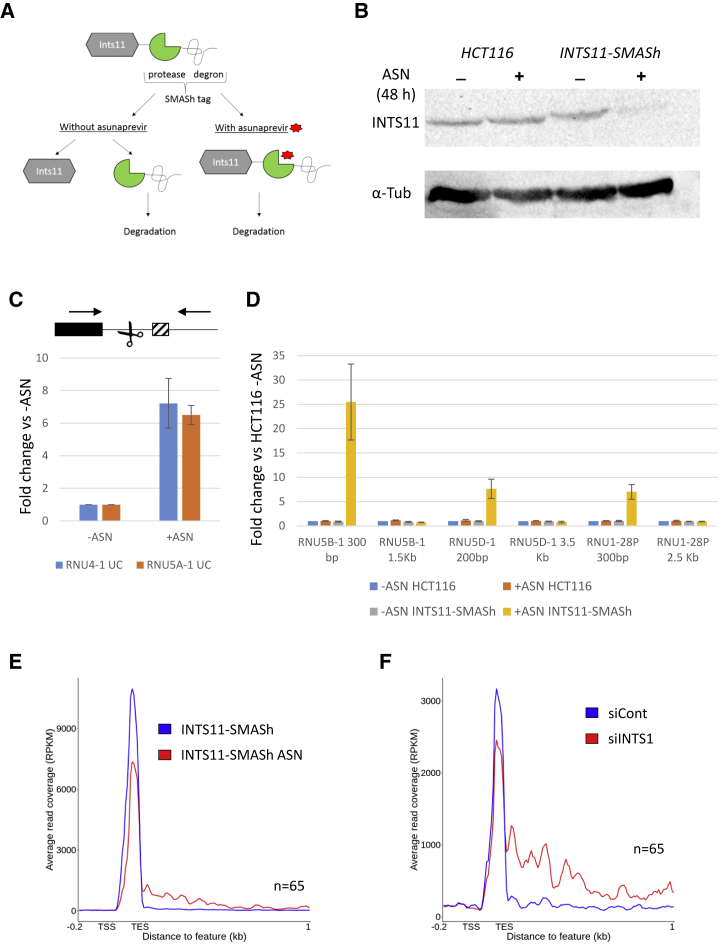
(A) Schematic of the SMASh tag system. Under normal conditions, the degron-encoded NS3 protease (green) removes the tag and the protein is stable. Addition of ASN (red) prevents this promoting proteosomal degradation.
(B) Western blot demonstrating ASN-dependent depletion of INTS11 after 48 h. Unmodified cells are shown as a control and the lower tubulin blot shows loading.
(C) qRT-PCR analysis of unprocessed (UC, uncleaved) RNU4-1 and RNU5A-1 transcripts in INTS11-SMASh cells treated or not with ASN. Levels in treated cells are shown relative to untreated cells after normalizing to spliced actin levels. The schematic shows snRNA (black rectangle), canonical Integrator cleavage site (scissors), downstream 3′ box (hatched box), and primers used (arrows). n = 4, error bars are standard error of the mean (SEM).
(D) qRT-PCR of RNU5 and RNU1 variants in HCT116 or INTS11-SMASh cells treated or not with ASN. Primers were used to detect read-through downstream of the annotated 3′ ends of each gene, with the distance indicated under each set of bars. Values are expressed as a fold change versus untreated HCT116 cells after normalizing to spliced actin levels. n = 3, error bars are SEM.
(E) Metagene plot of snRNA genes from nuclear RNA-seq performed on INTS11-SMASh cells treated or not with ASN. Signal is reads per kilobase of transcript, per million mapped reads (RPKM). TSS, transcription start site; TES, annotated snRNA 3′ end.
(F) Metagene plot of read-through at all expressed snRNA genes in chromatin-associated RNA-seq from HCT116 cells treated with control or INTS1-specific siRNAs. Signal is RPKM.