Figure 3.
Redundant RNA Cleavage and Direct Termination Generate Some RNA 3′ Ends at snRNA Genes
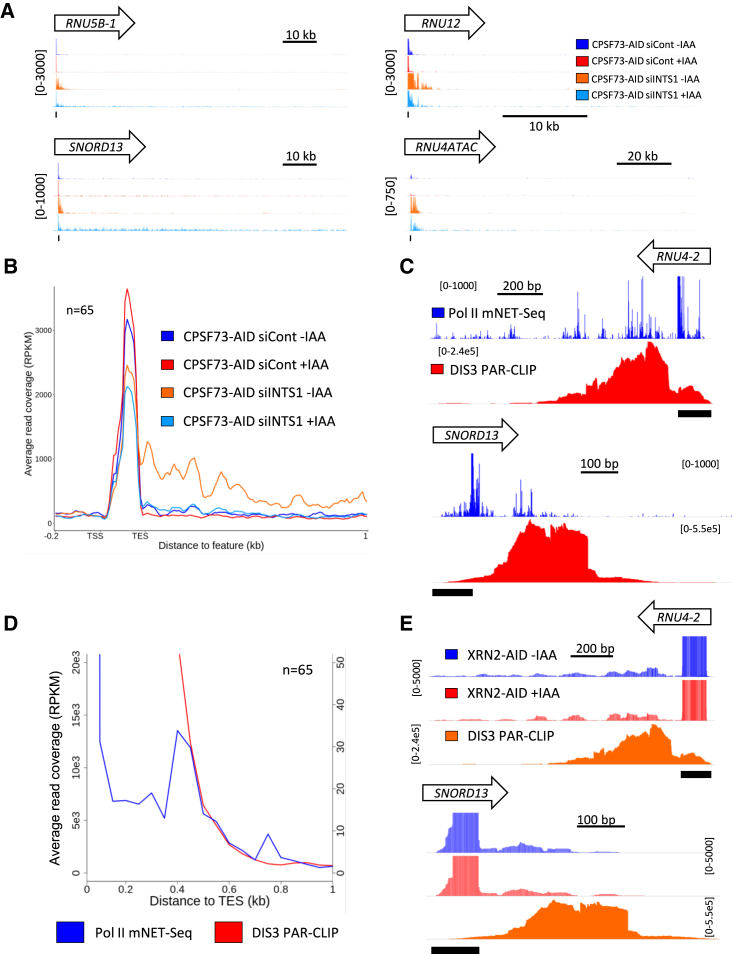
(A) Integrated Genomics Viewer (IGV) traces of RNU5B-1, SNORD13, RNU12, and RNU4ATAC (location indicated by black bar under each trace) from chromatin-associated RNA-seq performed in CPSF73-AID cells transfected with control or INTS1 siRNAs before treatment or not with auxin. The y axis shows RPKM.
(B) Metaplot analysis of all snRNAs derived from samples used in (A). The y axis scales are RPKM. The –IAA samples (siCont and siINTS1) are the same as those shown in Figure 1F, as these data were obtained in the same experiment.
(C) IGV tracks of RNU4-2 and SNORD13 (location indicated by black bar under each trace) comparing Pol II mNET-seq (blue) and published DIS3 PAR-CLIP (red) signals. mNET-seq signal is mapped to the 3′-most nucleotide of the read deriving from Pol II’s active center. “Spikes” of signal represent sites of greater Pol II occupancy. The y axis signals are RPKM.
(D) Metaplot of mNET-seq versus DIS3 PAR-CLIP (Szczepińska et al., 2015) at all expressed snRNAs. The Pol II mNET-seq signal is very high at the final nucleotide of snRNAs because of the detection of their incorporation into spliceosomes (Nojima et al., 2015). Therefore, a zoomed view is provided to highlight nascent signals beyond this region. The y axes show RPKM and signals are in 50-bp bins.
(E) IGV tracks of RNU4-2 and SNORD13 (location indicated by black bar under each trace) comparing DIS3 PAR-CLIP signals (Szczepińska et al., 2015) with nuclear RNA-seq obtained after auxin-dependent depletion of XRN2 (1-h depletion) (previously presented in Eaton et al., 2018). The y axis signals are RPKM.