Figure 4.
Analysis of Full-Length snRNA Precursors by Long-Read Sequencing
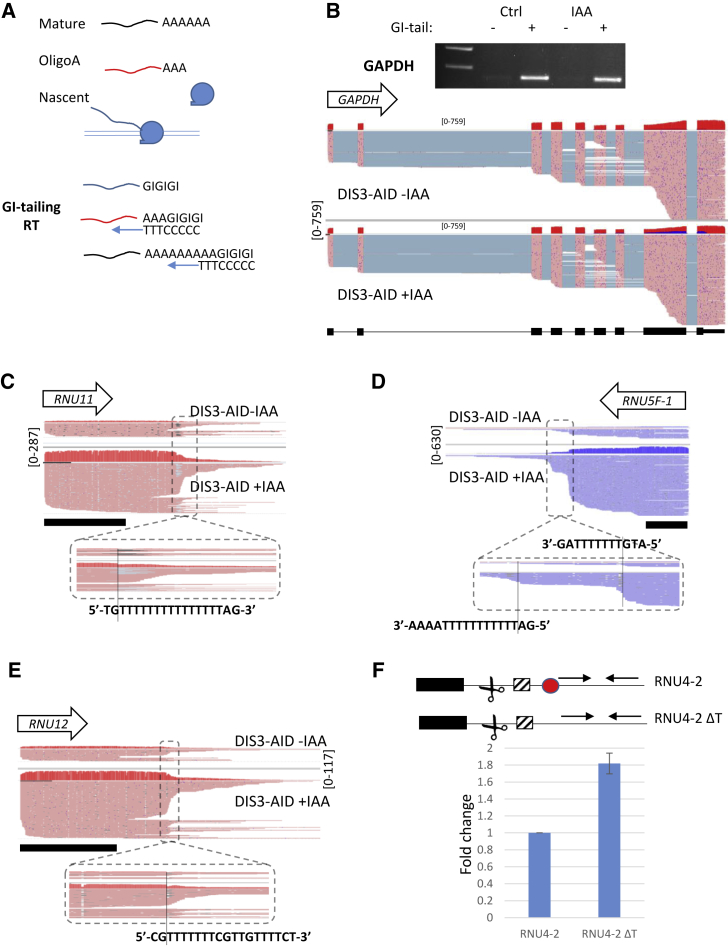
(A) Schematic of long-read sequencing strategy. Nuclei contain mature polyadenylated RNAs with long poly(A) tails (black), terminated oligoadenylated species that might be exosome substrates (red), and unadenylated (blue) transcripts that, for example, might derive from Pol II. GI-tailing allows the sequencing of RNAs with pre-existing poly(A) (black) or oligoA (red) tails following an oligo-dC(T)3-primed cDNA synthesis step.
(B) Agarose gel analysis of GAPDH transcripts isolated from DIS3-AID cells treated or not with auxin and/or GI-tailed, as indicated. Also shown is long-read sequencing tracks of GAPDH in GI-tailed samples. Each track shows coverage density at the top (red) scaled as transcripts per million (TPM) with individual reads displayed underneath. Individual reads show exons (pink) linked by introns removed by splicing. White space indicates unmapped regions between reads presumably truncated at their 5′/3′ ends. The blue signal coverage in auxin-treated samples derives from an annotated anti-sense RNA degraded by DIS3.
(C–E) Long-read sequencing tracks of RNU12, RNU11, and RNU5F-1 snRNAs from the GI-tailing experiment. Read density (TPM) and individual reads are shown. Regions of focused 3′ termini are indicated with a dashed box, and part of their primary sequence is shown. The black bar under each trace denotes the size and position of each mature snRNA as annotated.
(F) qRT-PCR from HCT116 cells transfected with a plasmid expressing RNU4-2 or a derivative lacking a downstream T-tract (red circle). Primers were used to detect read-through RNA beyond the T-tract, as shown in the schematic. Graph shows fold increase relative to the unmodified construct after normalizing to GFP levels from a co-transfected control plasmid. n = 5. Error bars are SEM.