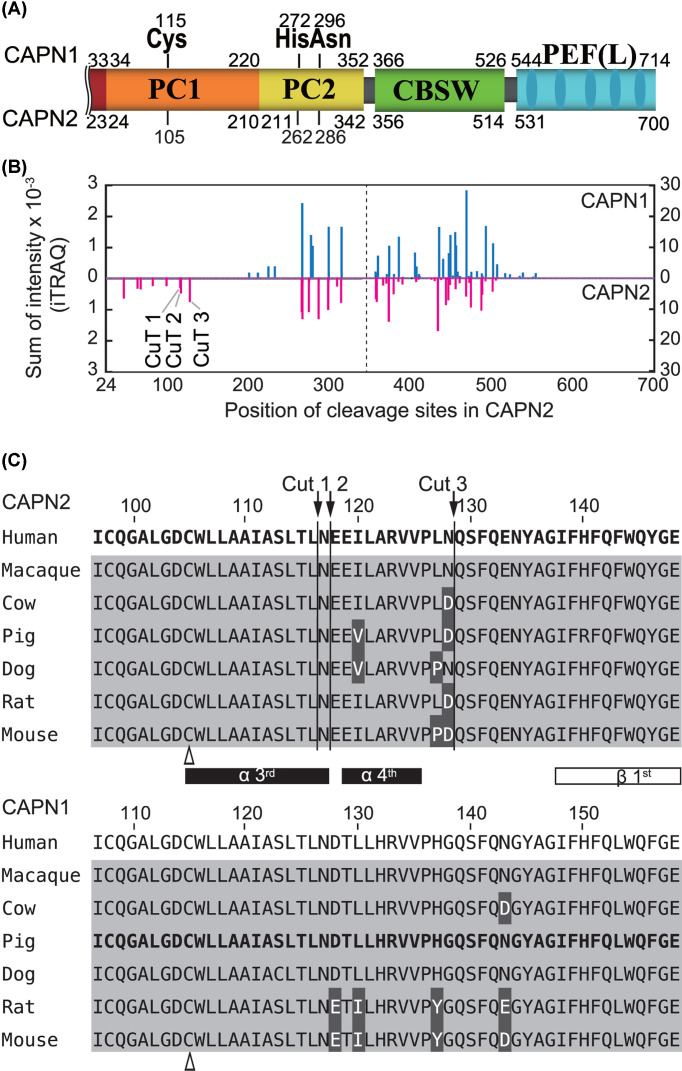
Figure 3. Autolysis of C1 and C2.
(A) Domain structure of CAPN1 and CAPN2. Due to its extended N-terminal sequence, CAPN1 (catalytic subunit of C1) is 14 amino acids longer than CAPN2 (catalytic subunit of C2). The alignment of the two amino acid sequences was adjusted using the catalytic amino acid residues as indices. (B) Comparison of autolytic sites in catalytic subunits, CAPN1 and CAPN2. Using the identified peptide sequence of C1 and C2 in the LC-MS/MS data described in Figure 2, autolytic sites of C1 and C2 are defined and the SI values were calculated as in Figure 2B (Supplementary Tables S2 and S3). C2-specific cleavage sites were identified in the PC1 domain of CAPN2, and were designated CuT1–3 (cleavage site unique to calpain-2 (Two)). Cleavage sites are located C-terminal side of the 116th, 117th and 128th amino acids. In the right half of the panel, i.e., beyond the dashed line, the scale is 10-fold larger than the left. (C) Amino acid sequence of mammalian CAPN1 and CAPN2 in PC1 domain. The amino acid sequence covering the P20 position from CuT1 to the P' 20 position from CuT3, 97–148 aa of CAPN2 is aligned with the corresponding sequences from mammalian orthologs (upper panel). The α-helices and β-strand are drawn according to the human ortholog [33]. The corresponding region of CAPN1 is also aligned accordingly (lower panel). Amino acid sequence identities in the selected region are 94.6% and 92.9% for CAPN2 and CAPN1, respectively. Sequences for human CAPN2 and pig CAPN1, the latter of which was actually used in the experiment, are in bold. Amino acid residues identical to and different from human sequence are highlighted in gray and black, respectively. Arrows, CuT1–3; open arrowhead, active site Cys residue.