Figure 1.
osbag4 Mutants Exhibit Salt Stress–Sensitive Phenotypes.
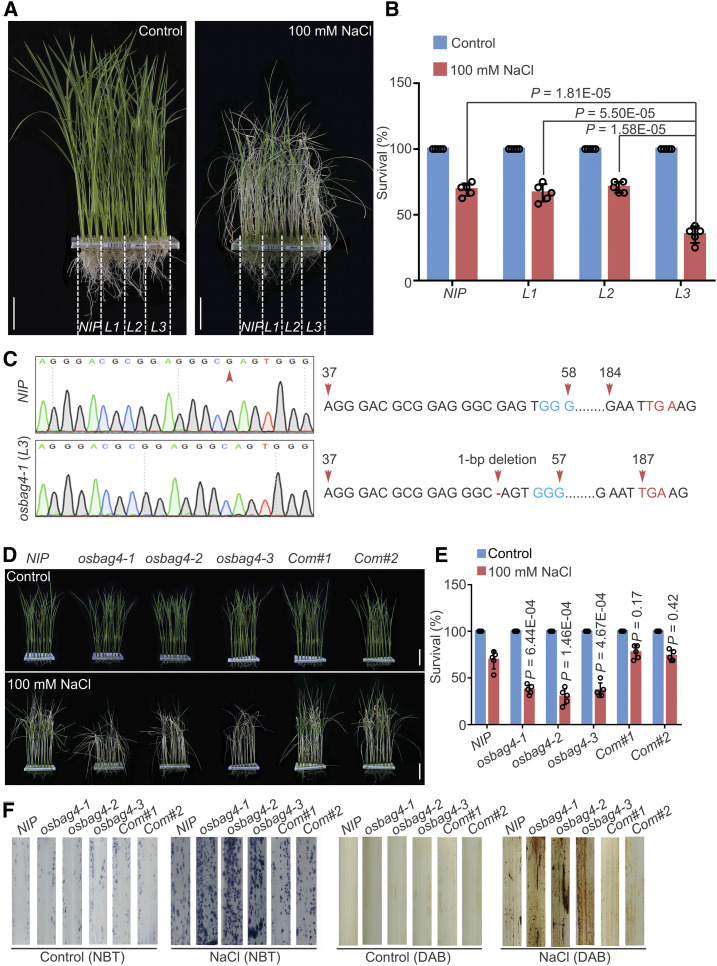
(A) and (B) Mutant lines obtained from the CRISPR/Cas9 mutant pool RGKO-ALL (L1, L2, and L3) were screened for salt stress sensitivity. NIP was used as a control. Images were captured (A), and survival rates were measured (B) before 100 mM NaCl treatment and after recovery from NaCl treatment Data in (B) represent means ± sd (n = 5, five biological experiments were performed with 24 plants in each). Individual values (black circles) are shown. Differences between NIP, L1, L2, and L3 were evaluated by Student’s t test. Bars in (A) = 4.0 cm.
(C) Sanger sequencing chromatography showing the wild-type and mutated forms of OsBAG4 in NIP and osbag4-1 (L3). The mutation in osbag4-1 includes a deletion of G leading to a premature stop codon.
(D) and (E) Images (D) and survival rates (E) of NIP, osbag4 mutants (osbag4-1, osbag4-2, and osbag4-3), and two OsBAG4 complementation lines (Com#1 and Com#2) before 100 mM NaCl treatment and after recovery from NaCl treatment. Data in (E) represent means ± sd (n = 5, five biological experiments were performed with 24 plants in each). Individual values (black circles) are shown. Statistical analyses were performed by comparing three independent osbag4 mutants and two OsBAG4 complementation lines with NIP plants by Student’s t test. Bars in (D) = 4.5 cm.
(F) ROS detection in the leaves of NIP, osbag4 mutants, and two OsBAG4 complementation lines under normal and salt stress conditions. Leaves stained with NBT and DAB were used to assess O2– and H2O2 accumulation, respectively. Seedlings were treated with or without 100 mM NaCl for 24 h treatment before staining.