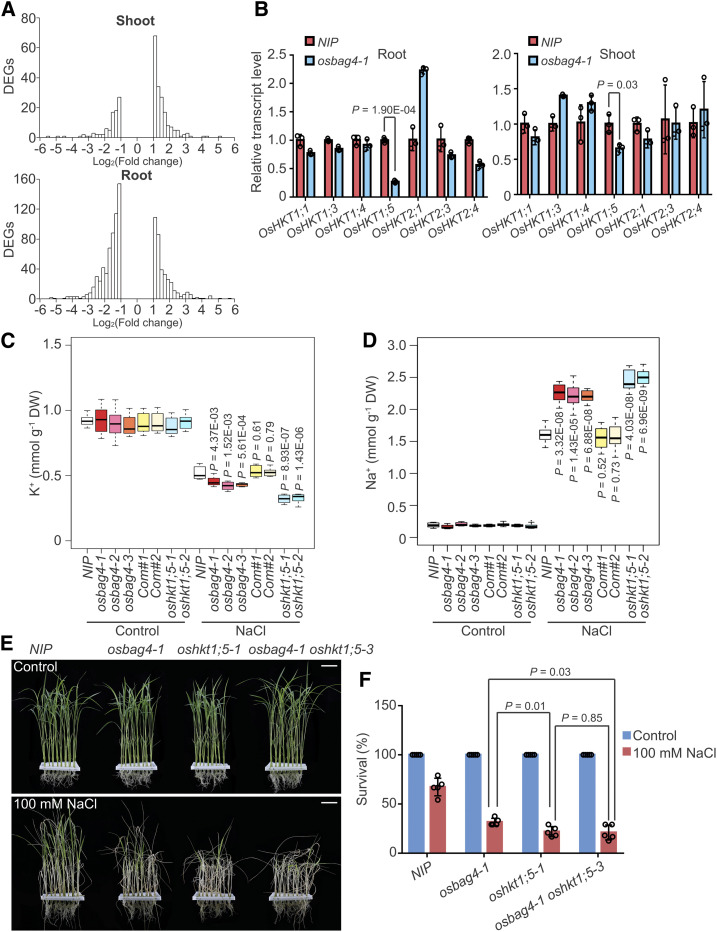
Figure 2.
OsHKT1;5 Acts Genetically Downstream of OsBAG4.
(A) Histograms of fold changes for up- and downregulated DEGs in shoot and root comparing osbag4-1 and NIP. “x axis” shows the bins of log2(Fold change) in RNA expression, and “y axis” is the number of DEGs whose log2(Fold change) expression fall in each bin.
(B) Expression levels of OsHKT family genes in the root or shoot of NIP and osbag4-1. Data represent means ± sd (n = 3, roots or shoots from five NIP or osbag4-1 seedlings were pooled and harvested for RNA extraction and RT-qPCR in each biological replicate). Statistical analyses were performed using Student’s t test. Individual values (black circle) are shown.
(C) and (D) Box plot showing K+ (C) and Na+ (D) contents in the shoots of the indicated genotypes. Four-week-old seedlings were treated with or without 100 mM NaCl for 5 d before measurement of ion contents (n = 8, eight plants of each genotype were used to measure Na+ and K+ levels). Differences between NIP and mutants or complementation lines were evaluated with Student’s t test. DW, dry weight.
(E) and (F) Images (E) and survival rates (F) of NIP, osbag4-1, oshkt1;5-1, and osbag4-1 oshkt1;5-3 double mutant before 100 mM NaCl treatment and after recovery from NaCl treatment. Data in (F) represent means ± sd (n = 5, five biological experiments were performed with 24 plants in each). Individual values (black circle) are shown. Statistical analyses were performed by Student’s t test. Bars in (E) = 3.2 cm.