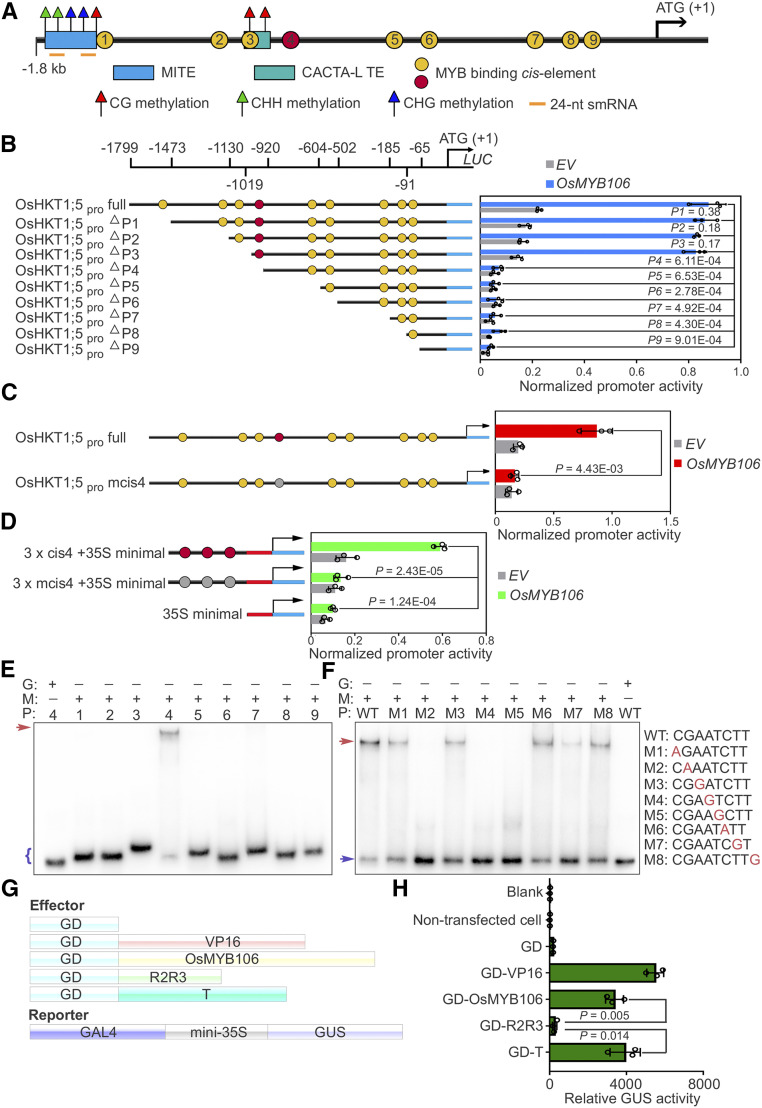
Figure 5.
OsMYB106 Activates OsHKT1;5 Expression by Binding to the cis-Element Located in the OsHKT1;5 Promoter.
(A) Schematic representation of the promoter structures of OsHKT1;5.
(B) Deletion analysis of OsHKT1;5 promoter activity. Left: schematic representation of a deletion series of the 1799-bp OsHKT1;5 promoter (OsHKT1;5pro full). The numbers displayed in the schematic of the OsHKT1;5 promoter indicate the distance from the OsHKT1;5 translation start site. Map indicates relative length and regions of 5′-deleted promoter versions cloned 5′ of a luciferase (Luc) coding sequence. Yellow and red circles indicate the MYBE represented in (A). Right, graph shows basal promoter activities measured in the presence of GFP (EV) and activities induced by FLAG-OsMYB106. Data are means ± sd (n = 3, transfection experiments were performed three times). Individual values (black circle) are shown. P-values were calculated versus the LUC activity driven by the full-length promoter activated by OsMYB106 (Student’s t test). Normalized promoter activity was calculated by normalizing LUC activity to GUS activity.
(C) Analysis of the 1799-bp OsHKT1;5 promoter (OsHKT1;5pro full) and 1799-bp OsHKT1;5 promoter containing a mutated form of the fourth cis-element (OsHKT1;5pro mcis4) in response to OsMYB106. Data represent means ± sd (n = 3, transfection experiments were performed three times). Individual values (black circles) are shown. Statistical analysis was performed by Student’s t test. Normalized promoter activity was calculated by normalizing LUC activity to GUS activity.
(D) Analysis of three tandem repeats of the fourth cis-element (cis4; red circles) and a mutated form of the fourth cis-element (mcis4; gray cycles) with the 35S minimal promoter in response to OsMYB106. Data represent means ± sd (n = 3, transfection experiments were performed three times). Individual values (black circles) are shown. Statistical analysis was performed by Student’s t test. Normalized promoter activity was calculated by normalizing LUC activity to GUS activity.
(E) Results of EMSA. Recombinant OsMYB106 protein bound to the consensus fourth cis-element located at OsHKT1;5 promoter. The red arrow denotes the shifted probe, and the purple parenthesis denotes the free probe. G, GST; M, GST-OsMYB106; P, probe (the numbers represent the predicted cis-element within the OsHKT1;5 promoter in [A]).
(F) Results of EMSA for single-nucleotide mutations of the eight nucleotides in the fourth cis-element. The red arrow denotes the shifted probe; the purple arrow denotes the free probe. WT, the wild-type consensus sequence identified in (E).
(G) and (H) Schematic diagrams of constructs (G) and quantification of GUS reporter activity (H) in NIP protoplasts cotransfected with effector constructs and GAL4-mini35Spro:GUS reporter. GUS activity was measured after protoplasts were incubated in the dark for 20 to 22 h. In (H), data represent means ± sd (n = 3, transfection experiments were performed three times). Individual values (black circle) are shown. Statistical analysis was performed by Student’s t test. Blank, empty well; VP16, herpes simplex virus VP16 activation domain.