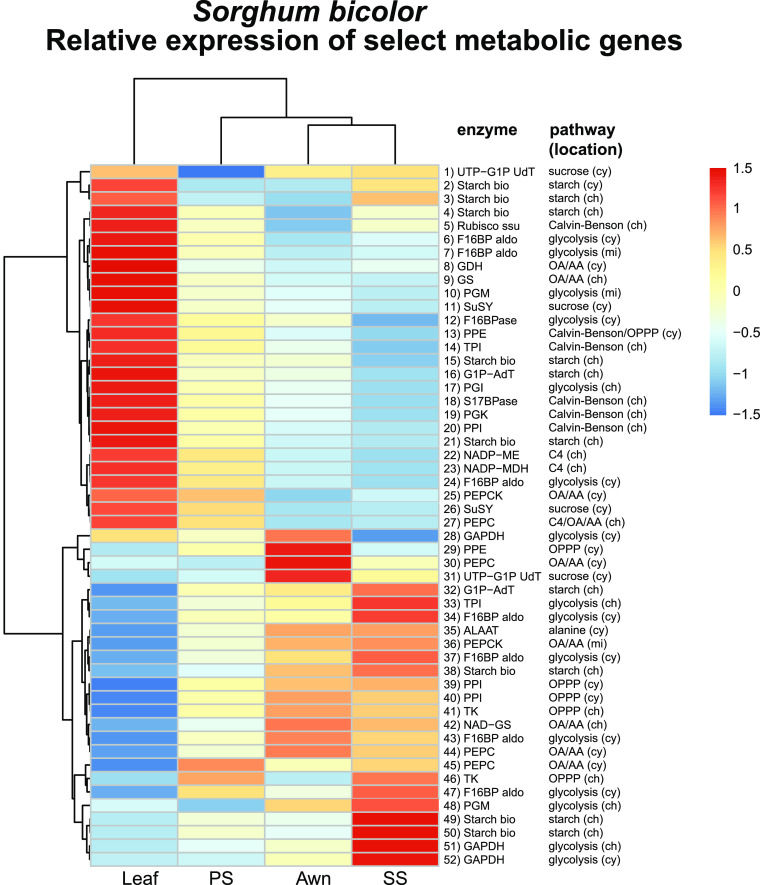
Figure 5.
S. bicolor.
Relative expression of genes encoding biosynthetic enzymes immediately responsible for producing the metabolites labeled with 13C, a subset extracted from full set of 922 DE metabolic genes in Supplemental Figure 3. Colors reflect scaled z-scores of log2-normalized expression values. Labels of genes indicate enzyme name, biochemical process, and subcellular localization: 1) UTP-G1P UdT, UTP-glucose-1-phosphate-uridyltransferase, Sobic.002G291200.1; 2) starch synthase, Sobic.001G239500.2; 3) starch synthase, Sobic.010G047700.1; 4) starch synthase, Sobic.002G116000.1; 5) ribulose-1,5-bisphosphate carboxylase/oxygenase, small subunit, Sobic.005G042000.1; 6) F16BP aldo, fructose-1,6-bis-phosphate aldolase, Sobic.005G056400.1; 7) F16BP aldo, fructose-1,6-bis-phosphate aldolase, Sobic.008G053200.1; 8) GDH, glutamate dehydrogenase, Sobic.003G188400.1; 9) GS, glutamine synthetase, Sobic.006G249400.1; 10) PGM, phosphoglucomutase, Sobic.003G222500.1; 11) SuSY, sucrose phosphate synthase, Sobic.004G068400.1; 12) F16BPase, fructose-1,6-bisphosphatase, Sobic.003G367500.1; 13) PPE, phosphopentose epimerase, Sobic.001G491000.1; 14) TPI, triose phosphate isomerase, Sobic.002G277100.1; 15) starch synthase, Sobic.006G221000.1; 16) G1P-AdT, glucose-1-phosphate adenyltransferase, Sobic.007G101500.1; 17) PGI, phosphoglucoisomerase, Sobic.002G230600.1; 18) S17BPase, sedoheptulose 1,7-bisphosphatase, Sobic.003G359100.1; 19) PGK, phosphoglycerate kinase, Sobic.009G183700.1; 20) PPI, phosphopentose isomerase, Sobic.001G069000.1; 21) starch synthase, Sobic.004G238600.1; 22) NADP-ME, NADP-malic enzyme, Sobic.003G036200.1; 23) NADP-MDH, NADP-malate dehydrogenase, Sobic.007G166300.1; 24) F16BPase, fructose-1,6-bisphosphatase, Sobic.010G188300.1; 25) PEPCK, phosphoenol pyruvate carboxykinase, Sobic.004G338000.1; 26) SuSY, sucrose phosphate synthase, Sobic.003G403300.1; 27) PEPC, phosophoenol pyruvate carboxylase, Sobic.010G160700.1; 28) GAPDH, glyceraldehyde-3-phosphate dehydrogenase, Sobic.005G159000.1; 29) PPE, phosphopentose epimerase, Sobic.002G257300.1; 30) PEPC, phosphoenol pyruvate carboxylase, Sobic.004G106900.1; 31) UTP-G1P UdT, Sobic.006G213100.1; 32) G1P-AdT, glucose-1-phosphate adenyltransferase, Sobic.002G160400.1; 33) TPI, triose phosphate isomerase, Sobic.003G072300.2; 34) F16BP aldo, fructose-1,6-bisphosphate aldolase, Sobic.004G146000.1; 35) ALAAT, alanine amino transferase, Sobic.001G260701.1; 36) PEPCK, phosphoenol pyruvate carboxykinase, Sobic.006G198400.2; 37) F16BP aldo, fructose-1,6-bisphosphate aldolase, Sobic.003G393900.1; 38) starch synthase, Sobic.007G068200.1; 39) PPI, phosphopentose isomerase, Sobic.003G182400.1; 40) PPI, phosphopentose isomerase, Sobic.008G135701.1; 41) TK, transketolase, Sobic.010G024000.2; 42) NAD-GS, Sobic.003G258800.1; 43) F16BP aldo, fructose-1,6-bisphosphate aldolase, Sobic.003G096000.2; 44) PEPC, phosphoenol pyruvate carboxylase, Sobic.003G301800.1; 45) PEPC, phosphoenol pyruvate carboxylase, Sobic.002G167000.1; 46) TK, transketolase_Sobic.009G062800.1; 47) F16BP aldo, fructose-1,6-bisphosphate aldolase, Sobic.009G242700.1; 48) PGM, phosphoglucomutase, Sobic.001G116500.1; 49) starch synthase, Sobic.010G022600.1; 50) starch synthase, Sobic.010G093400.1; 51) GAPDH, glyceraldehyde-3-phosphate dehydrogenase, Sobic.004G056400.1; 52) GAPDH, glyceraldehyde-3-phosphate dehydrogenase, Sobic.004G205100.1. cy, cytosolic localized; ch, chloroplast localized; mi, mitochondrial localized; OA/AA, organic acid/amino acid metabolism.