Abstract
Osteosarcoma is a severe malignant tumor. Several studies indicated that lncRNA prostate cancer-associated transcript 6 (PCAT6) promoted the development of multiple types of cancers. Studies have also revealed that MDM2 could aggravate tumor symptoms inhibiting P53 expression. However, whether lncRNA PCAT6 could affect the proliferation and metastasis of osteosarcoma cells by regulating P53 expression is unclear. The present study established lncRNA PCAT6-overexpressing osteosarcoma cells. Cell Counting Kit-8, wound healing and Transwell assays were performed to detect the change in proliferation, migration and invasion of these cells, respectively. Subsequently, E3 ubiquitin-protein ligase Mdm2 (MDM2), P53 and P21 expression were determined using western blotting. Finally, MDM2 expression was inhibited and the proliferation, migration and invasion of these cells was determined again. The present study found that the proliferation, migration and invasion of osteosarcoma cells increased following overexpression of lncRNA PCAT6. MDM2 expression was upregulated while the levels of P53 and P21 decreased following overexpression of lncRNA PCAT6. However, the proliferation, migration and invasion of osteosarcoma cells were inhibited following MDM2 knockdown. Additionally, P53 and P21 was rescued following MDM2 knockdown. To conclude, lncRNA PCAT6 promoted the proliferation, migration and invasion of osteosarcoma cells by promoting the expression of MDM2 and suppressing the expression of P53 and P21.
Keywords: osteosarcoma, long non-coding RNA prostate cancer-associated transcript 6, E3 ubiquitin-protein ligase Mdm2, P53, proliferation
Introduction
Osteosarcoma is a common malignant bone tumor, which is highly lethal to adolescents and the elderly (1,2). Osteosarcoma cells have strong metastatic and invasion abilities, the disease could be aggravated by lung metastases at early stages (3). Although numerous studies in recent years have provided diverse treatment options, including gene therapy targeting long non-coding RNA (lncRNA) or microRNA (miR/miRNA) for osteosarcoma, the disease is still not fully understood (4,5).
Long non-coding RNA (lncRNA) is a type of the non-coding RNA molecule composed of >200 nucleotides (6,7). A study showed that lncRNAs play a critical regulatory role in the proliferation, differentiation and apoptosis of osteosarcoma cells (8). Several studies also suggested that lncRNA expression could regulate the proliferation, migration and invasion of multiple types of malignant tumor cells (9,10). LncRNA prostate cancer-associated transcript 6 (lncRNA PCAT6) promoted the proliferation of prostate cancer cells in an androgen-independent manner (11). Furthermore, high levels of lncRNA PCAT6 also promoted the proliferation, migration and invasion of non-small cell lung cancer cells and ovarian cancer (12,13). LncRNA PCAT6 was reported to act as a miRNA sponge to regulate the proliferation, migration and invasion of osteosarcoma (14). However, the effect of lncRNA PCAT6 on the occurrence of osteosarcoma remains to be elucidated.
P53 is a classic tumor suppressor gene (15). Higher levels of P53 expression could inhibit the proliferation and metastasis of breast, liver and lung cancer cells (15). P21, also known as CDKN1A, plays a crucial role in cell cycle progression (16). P21 could also suppress the proliferation of multiple cancer cells by inducing cell cycle arrest (17). A study revealed that P53 could activate the expression of P21 and therefore enhance the antitumor effects of P53/P21 (18). Another study indicated that MDM2 could promote the occurrence and development of malignant tumors by inducing P53 ubiquitination (19). Several studies also indicated that E3 ubiquitin-protein ligase Mdm2 (MDM2), which negatively regulates p53 protein, could promote the development of ovarian (20) and lung cancer (21). However, whether lncRNA PCAT6 could promote the development of osteosarcoma by promoting the expression of MDM2 is unclear.
The present study established lncRNA PCAT6- overexpressing osteosarcoma cell lines. Subsequently, the proliferation, migration and invasion of these cells were determined with Cell Counting Kit (CCK)-8, wound healing and Transwell assays, respectively. MDM2 expression was then suppressed in the lncRNA PCAT6-overexpressing osteosarcoma cell line by transfection. Following which, the proliferation and metastasis of these cells were detected again. The results of these assays could reveal the effect of lncRNA PCAT6 on osteosarcoma development.
Materials and methods
Cell culture and transfection
Normal osteoblasts (hFOB1.19) and human osteosarcoma cell lines (Saos2, MG63, U2OS and HOS) used in the present study were purchased from American Type Culture Collection. Cells were cultured with RPMI-1640 medium (HyClone; Cytiva) supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.). Cells were cultured in 37°C humid atmosphere with 5% CO2. LncRNA PCAT6 overexpression lentiviruses (4 µg/ml; 1 ml) were designed and purchased from Shanghai GeneChem Co., Ltd. 293T cells (American Type Culture Collection) were used to package lncRNA PCAT6 overexpression lentivirus vectors or empty lentivirus vectors (NC groups). The MG63 cells in the control group were not treated with lentiviruses and transfection reagent. Small interfering RNA (si)-MDM2 (si-MDM2-1, 5′CAGCCATCAACTTCTAGTA3′; si-MDM2-2, 5′CCACCTCACAGATTCCAGCTTCAAGAGAGCTGGAATCTGTGAGGTGG-3′) lentivirus (4 µg/ml; 1 ml) and corresponding vector were also obtained from Shanghai GeneChem Co., Ltd. Polybrene (Shanghai GeneChem Co., Ltd.) was used to increase transfection efficacy. After 24 h of transfection, MG63 cells were used for further experiments.
Animal assays
A total of 30 BALB/c nude mice (male; 4-week-old; 18–20 g) were obtained from the Shanghai Lingchang Biotechnology Company (Shanghai, China). Mice were raised in a sterile laminar flow cabinet with a 12-h light/dark cycle at 25±1°C and 40–60% humidity and free access to food and water.
Subsequently, MG63 cells (1×106) were injected into the thigh joint of nude mice by subcutaneous injection. After two weeks, these mice were anesthetized with ketamine (80 mg/kg) and xylazine (7 mg/kg). The mice were then placed in a carbon dioxide (CO2) euthanasia chamber (Shanghai Yuyan Instruments Co., Ltd.) and sacrificed by excess CO2. The controlled flow rate of CO2 was 20% of the volume of the euthanasia chamber per minute. Once the animal lost consciousness, the flow rate was increased to 100% of the euthanasia chamber volume per minute. Mice tumors were collected for subsequent experimentation. The percentage of weight loss was also calculated using the following formula: (Daily weight-weight on the first day)/weight on the first day ×100%. The weight and tumor volume of mice were recorded each day. Tumor weight was detected with an electronic balance. The formula for calculating tumor volume of mice is as follows: Tumor volume=0.5 × length × width2. Cells transfected with pc-DNA PCAT6 or si-MDM2 were injected into the back of nude mice (1×106 cells). The mice were divided into four groups (n=7 per group): pcDNA-negative control (NC), pcDNA-PCAT6, pcDNA-PCAT6 + si-MDM2 and pcDNA-PCAT6 + RG7112 (5 µm; cat. no. HY-10959; MedChemExpress).
Two mice died during the experiments and were not used to perform the analysis of the result. The possible causes were as follows: i) The rapidly growing tumor led to the restricted movement of the mice's forelimbs, which affected feeding and led to death; or ii) the influences of the experimental environment. All animal experiments were performed in accordance with the animal experimental guidelines set by the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The study was approved by the Experimental Animal Ethical Committee of Zhejiang Hospital (approval no. JUMC2019-019).
CCK-8 assay
MG63 cells were plated into a 96-well plate (2×103 cells/well). Cell proliferation abilities were detected using a CCK-8 assay (Dojindo Molecular Technologies, Inc.) and performed according to the manufacturer's instructions. After 24 h, 10 µl CCK-8 solution (Dojindo Molecular Technologies, Inc.) was diluted in culture medium and added into the 96-well plates. Subsequently, cells were incubated at 37°C for 1 h. The absorbance of the cells at a wavelength of 450 nm was detected with a spectrophotometer (Thermo Fisher Scientific, Inc.). The other two plates were detected after 48 and 72 h of culture.
Wound healing assay
Prior to experimentation, cells of different groups were plated into six-well plates (4×105 cells/well). Once adhered, cells were cultured in serum-free medium for 12 h. A scratch was then created with a pipette tip. The width of the scratch was photographed and recorded at 0 h (magnification, ×100). Subsequently, the width of the scratch was photographed after 24 h (magnification, ×100) with a light microscope (Olympus Corporation). The migration rates were calculated with the following formula: Migration rate=[(width at 0 h-width at 24 h)/width of 0 h] ×100%.
Transwell assay
Cells were cultured in serum-free medium for 12 h before experimentation. Cells (1×105/ml) were then plated into Transwell chambers (Corning Inc.). Complete medium with 10% FBS was also added to the lower Transwell chamber. Following 24 h, the cells in the opposite basement membrane of the Transwell chamber was fixed with 4% paraformaldehyde for 10 min and then stained with crystal violet for 5 min at room temperature (Thermo Fisher Scientific, Inc.). Stained cells were calculated under a light microscope (magnification, ×100; Olympus Corporation).
Hematoxylin and eosin (HE) staining
Tumor tissues were collected and used for experiments after mice euthanasia. After tumor tissues were fixed in 4% paraformaldehyde for 48 h at 4°C, tumor tissues were embedded with paraffin and then cut into 5-µm sections. Slices were subsequently stained with hematoxylin and eosin (Thermo Fisher Scientific, Inc.) for 5 min at room temperature. Three slices were observed and photographed under a light microscope (magnification, ×200; Olympus Corporation).
Immunohistochemistry
Tumor tissues were fixed in 4% paraformaldehyde for 48 h at 4°C. Subsequently, tumor tissues were dehydrated and cut into 5-µm sections. An appropriate amount of normal goat serum (Beyotime Institute of Biotechnology) was added to the slide tissue for non-specific antigen blocking for 30 min at 37°C. Subsequently, Ki-67 expression in tumor tissues was detected with Ki-67 antibody (1:5,000; cat. no. ab15580; Abcam) according to the manufacturer's instructions. Following which, goat anti-rabbit biotinylated secondary antibody (1:40,000; cat. no. ab7089; Abcam) was added to detect the primary antibody. Neutral balsam was used to mount the sections. The staining results were observed under a light microscope (magnification, ×200; Olympus Corporation).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was reverse transcribed into cDNA using a reverse transcription kit (cat. no. 4897030001; Roche Diagnostics) according to the manufacturer's instructions. SYBR Green (Thermo Fisher Scientific, Inc.) was used as the fluorescence indicator during the cDNA amplification process. The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 30 sec; followed by 40 cycles of denaturation at 95°C for 5 sec and annealing and extension at 60°C for 30 sec. The expression of targeted genes was calculated using the 2−∆∆Cq method (22). The following primer pairs were used for the qPCR: MDM2 forward, 5′-AGTAGCAGTGAATCTACAGGGA-3′ and reverse, 5′-CTGATCCAACCAATCACCTGAAT-3′; lncRNA PCAT6 forward, 5′-CCCCTCCTTACTCTTGGACAAC-3′ and reverse, 5′-GACCGAATGAGGATGGAGACAC-3′ and GAPDH forward, 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse, 5′-AGGGGCCATCCACAGTCTTC-3′.
Western blotting
Total protein (20 µg) was extracted from cells or tumor tissues using RIPA buffer (Beyotime Institute of Biotechnology). Protein concentration was determined using a BCA assay. Proteins were separated via 12% SDS-PAGE (Beyotime Institute of Biotechnology) and transferred to PVDF membranes (EMD Millipore). Subsequently, membranes were blocked in 5% skim milk powder solution at room temperature for 1 h and incubated with primary antibodies overnight at 4°C. The primary antibodies used were as follows: MMP-2 (1:1,000; cat. no. ab97779; Abcam), MMP-9 (1:1,000; cat. no. ab38898; Abcam), P21 (1:5,000; cat. no. ab109520; Abcam), P53 cat. no. (1:5,000; ab26; Abcam), MDM2 (1:1,000; cat. no. ab16895; Abcam) and GAPDH (1:5,000; cat. no. ab9485; Abcam). Subsequently, membranes were incubated with HRP-conjugated secondary antibodies (1:10,000; cat. nos. ab97040 and ab97080; Abcam) at room temperature for 1 h. On the second day, membranes were washed with PBS-Tween-20 (0.05%) three times. Finally, the bands were visualized with chemiluminescence reagent (EMD Millipore), followed by the analysis of gray value of protein bands using ImageJ software (version 1.46; National Institutes of Health).
Statistical analysis
Data were analyzed using GraphPad Prism 8.0 (GraphPad Software, Inc.). One-way ANOVA was used for comparison among multiple groups, followed by Tukey's post hoc test. Data were expressed as the mean ± SD. All experiments in were repeated three times. P<0.05 was considered to indicate a statistically significant difference.
Results
Overexpression of lncRNA PCAT6 promotes the proliferation of osteosarcoma cells
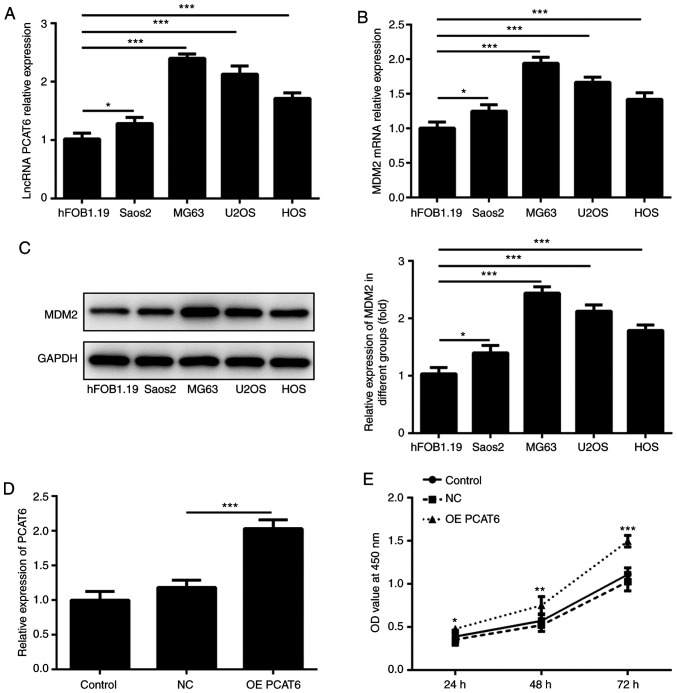
LncRNA PCAT6 was reported to be upregulated in osteosarcoma tissue (14). The present study detected the expression of lncRNA PCAT6 and MDM2 in osteosarcoma cell lines (hFOB1.19, Saos2, MG63, U2OS and HOS). As shown in Fig. 1A-C, lncRNA PCAT6 and MDM2 expression was highest in MG63 cells. Therefore, MG63 cells was selected for subsequent experiments. LncRNA PCAT6-overexpressing osteosarcoma cells were then established. Fig. 1D shows that lncRNA PCAT6 expression in the overexpression group was significantly higher compared with the NC group. CCK-8 assays were performed to detect the change in MG63 cell proliferation. Compared with the NC group, the proliferation of MG63 cells significantly increased following overexpression of lncRNA PCAT6 at 24, 48 and 72 h (Fig. 1E).
Figure 1.
LncRNA PCAT6 promotes osteosarcoma cell proliferation. (A) LncRNA PCAT6 expression in different cells was determined *P<0.05 and ***P<0.001. MDM2 expression in different cells was detected using (B) RT-qPCR and (C) western blotting *P<0.05 and ***P<0.001. (D) LncRNA PCAT6 expression was determined using RT-qPCR following lncRNA PCAT6 expression ***P<0.001. (E) Cell Counting Kit-8 assays were performed to detect osteosarcoma cell proliferation following lncRNA PCAT6 overexpression. *P<0.05, **P<0.01 and ***P<0.001 vs. NC. lncRNA PCAT6, long non-coding RNA prostate cancer-associated transcript 6; MDM2, E3 ubiquitin-protein ligase Mdm2; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; OE, overexpression; OD, optical density.
Overexpression of lncRNA PCAT6 enhances the migration and invasion of osteosarcoma cells
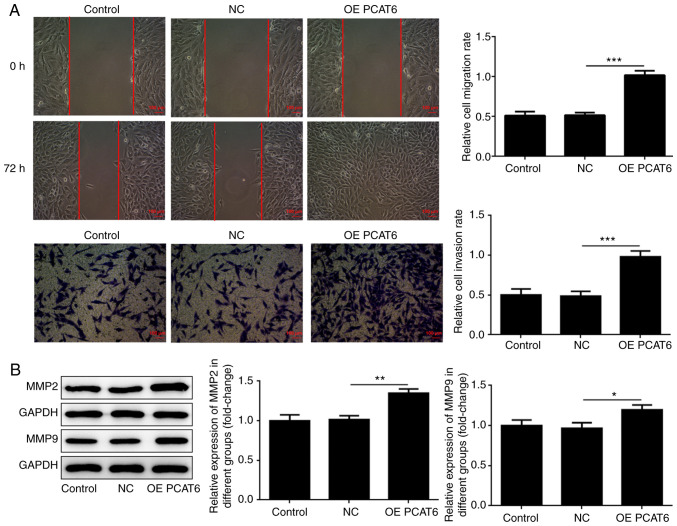
Wound healing and Transwell assays were performed to detect the migration and invasion abilities of MG63 cells following overexpression of lncRNA PCAT6. Based on the results (Fig. 2A), it was found that the migration and invasion of MG63 cells significantly increased following overexpression of lncRNA PCAT6 compared with the NC group. Increased levels of MMP-2 and MMP-9 were closely correlated with metastases of osteosarcoma (23,24). Subsequently, MMP-2 and MMP-9 expression was detected via western blotting. As shown in Fig. 2B, MMP-2 and MMP-9 expression in MG63 cells significantly increased following overexpression of lncRNA PCAT6 compared with the NC group.
Figure 2.
LncRNA PCAT6 enhances the migration and invasion of osteosarcoma cells. (A) The migration and invasion of osteosarcoma cells increased following overexpression of lncRNA PCAT6. (B) MMP-2 and MMP-9 expression was detected using western blotting. *P<0.05, **P<0.01 and ***P<0.001. lncRNA PCAT6, long non-coding RNA prostate cancer-associated transcript 6; NC, negative control; OE, overexpression.
Overexpression of lncRNA PCAT6 promotes the proliferation of osteosarcoma cells in vivo
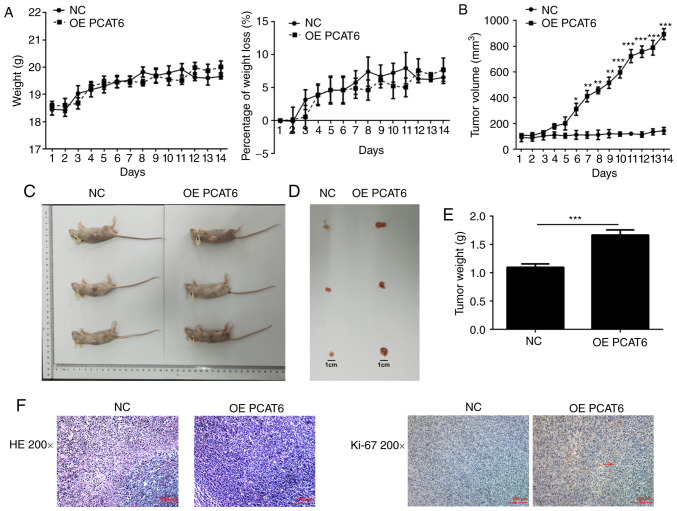
MG63 cells (negative control and overexpression group) were injected into mice by subcutaneous injection. The weight of these mice was measured each day for 14 days after injection. There was no significant difference in the weight and weight loss of mice between the PCAT6 overexpression group and control group (Fig. 3A). The tumor volume was significantly increased to varying degrees following overexpression of lncRNA PCAT6 (Fig. 3B). The maximum tumor diameter measured was 0.94 cm. The tumor weight significantly increased following overexpression of lncRNA PCAT6 compared with the NC group (Fig. 3C-E). Additionally, the maximum percentage of tumor weight out of total animal body weight was 8.59%. Tumor nuclei were heavily stained and the atypia was obvious using HE staining in the OE PCAT6 group compared with the NC group (Fig. 3F). Immunohistochemistry results (Fig. 3F) also showed that ki-67 expression was enhanced in tumor tissues overexpressing lncRNA PCAT6.
Figure 3.
Overexpression of lncRNA PCAT6 promoted the proliferation of osteosarcoma cells in vivo. (A) The daily weight of mice was recorded, and the percentage of weight loss was calculated. (B) Tumor volume. (C) Tumor size. (D) Tumor diameter. (E) Tumor weight. (F) HE staining in osteosarcoma tissue was performed to analyze pathological characteristics and immunohistochemistry was performed to detect ki-67 expression. *P<0.05, **P<0.01 and ***P<0.001 vs. NC. lncRNA PCAT6, long non-coding RNA prostate cancer-associated transcript 6; NC, negative control; OE, overexpression; HE, hematoxylin-eosin.
Overexpression of lncRNA PCAT6 promotes MDM2 expression
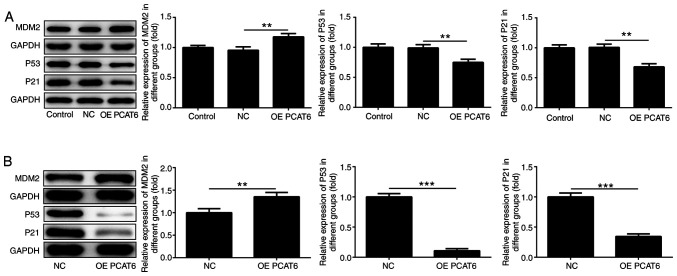
Subsequently, the expression of MDM2, P53 and P21 in MG63 cells and tumors was detected. The results showed that MDM2 expression significantly increased following lncRNA PCAT6 overexpression compared with the NC group (Fig. 4A and B). However, the levels of P53 and P21 significantly decreased following overexpression of lncRNA PCAT6 compared with the NC group.
Figure 4.
Overexpression of lncRNA PCAT6 promotes MDM2 expression. The expression of MDM2, P21 and P53 in (A) MG63 cells and (B) tumor tissues was determined using western blotting. **P<0.01 and ***P<0.001. lncRNA PCAT6, long non-coding RNA prostate cancer-associated transcript 6; NC, negative control; OE, overexpression; MDM2, E3 ubiquitin-protein ligase Mdm2.
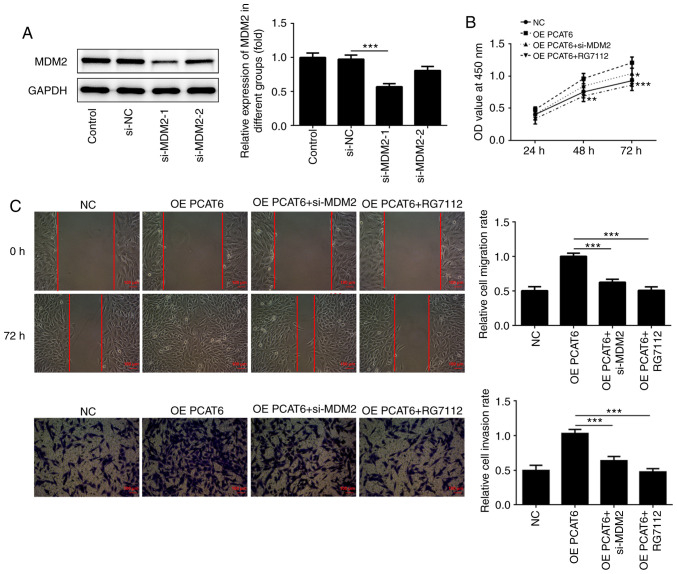
MDM2 knockdown weakens the proliferation, migration and invasion of osteosarcoma cells
Lentiviruses were designed to establish MDM2-knockdown osteosarcoma cells. As shown in Fig. 5A, MDM2 expression was inhibited following transfection with si-MDM2. However, the inhibition efficiency of si-MDM2-1 was higher compared with si-MDM2-2. Therefore, si-MDM2-1 was selected for subsequent experiments. Next, CCK-8 assays were performed to MG63 cell proliferation. The results showed that MG63 cell proliferation was suppressed following MDM2 knockdown compared with the OE PCAT6 group at 48 and 72 h (Fig. 5B). Similarly, compared with the OE PCAT6 group, the proliferation of these cells was also promoted after treatment with RG7112, an MDM2 inhibitor. The migration and invasion of MG63 cells were also significantly inhibited after MDM2 knockdown and RG7112 treatment compared with PCAT6 overexpression (Fig. 5C).
Figure 5.
Knockdown of MDM2 inhibits the proliferation, migration and invasion of osteosarcoma cells. (A) MDM2 expression was detected using western blotting. ***P<0.001. (B) Cell Counting Kit-8 assays were performed to detect osteosarcoma cell proliferation. *P<0.05, **P<0.01 and ***P<0.001 vs. OE PCAT6. (C) Wound healing and Transwell assays were performed to determine the migration and invasion of osteosarcoma cells following MDM2 knockdown, respectively. ***P<0.001. lncRNA PCAT6, long non-coding RNA prostate cancer-associated transcript 6; NC, negative control; OE, overexpression; MDM2, E3 ubiquitin-protein ligase Mdm2; si, small interfering RNA; OD, optical density.
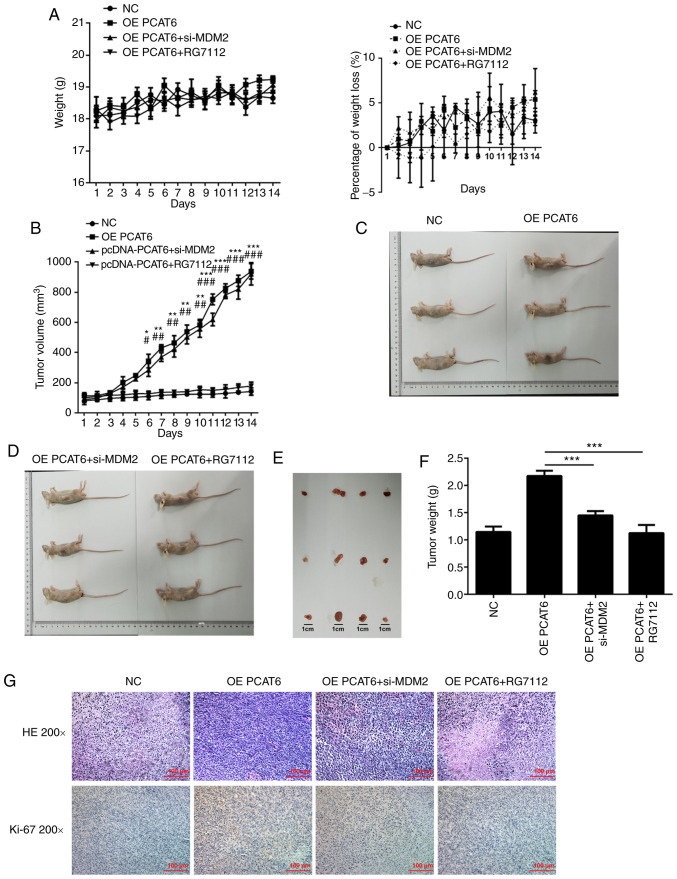
MDM2 knockdown suppresses the proliferation of osteosarcoma cells in vivo
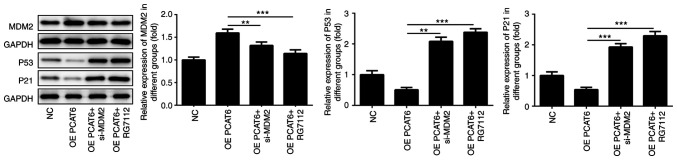
For these experiments, MG63 cells were injected into nude mice by subcutaneous injection. There was no significant difference in weight and weight loss between the mice of different groups (Fig. 6A). The tumor volume and tumor weight declined following MDM2 knockdown and RG7112 treatment (Fig. 6B and F). The mice and tumors in different groups are shown in Fig. 6C-E. The maximum percentage of tumor weight, out of total animal body weight, was 11.65% and the maximum tumor diameter measured was 0.98 cm. Compared with the OE PCAT6 group, the nucleolus partially disappeared while the nucleocytoplasmic ratio decreased in OE PCAT5+si-MDM2 and OE PCAT5+RG7112 groups through HE staining and immunohistochemistry results also showed that ki-67 expression was inhibited following MDM2 knockdown and RG7112 treatment (Fig. 6G). In previous studies, we found that lncRNA PCAT6 knockdown enhanced the expression of P53 (25). Therefore, the expression of P53 and P21 after MDM2 knockdown was detected. Knockdown of MDM2 rescued the expression of P21 and P53 (Fig. 7). MDM2 knockdown significantly increased p53 and p21 expression compared with the OE PCAT6 group, the effects of which were similar to RG7112 treatment.
Figure 6.
Knockdown of MDM2 suppresses osteosarcoma cell proliferation in vivo. (A) Mice weight. (B) Daily tumor volume. *P<0.05, **P<0.01 and ***P<0.001, OE PCAT6 group vs. OE PCAT6 + MDM2 group. #P<0.05, ##P<0.01 and ###P<0.001 OE PCAT6 group vs. OE PCAT6 + RG7112 group. (C-E) Tumor weight. (G and F) HE staining in osteosarcoma tissue and immunohistochemistry was performed to determine ki-67 expression in tumor tissues. ***P<0.001. lncRNA PCAT6, long non-coding RNA prostate cancer-associated transcript 6; NC, negative control; OE, overexpression; MDM2, E3 ubiquitin-protein ligase Mdm2; si, small interfering RNA.
Figure 7.
Knockdown of MDM2 rescues P53 and P21 expression. MDM2, P53 and P21 expression was detected following MDM2 knockdown. **P<0.01 and ***P<0.001. lncRNA PCAT6, long non-coding RNA prostate cancer-associated transcript 6; NC, negative control; OE, overexpression; MDM2, E3 ubiquitin-protein ligase Mdm2; si, small interfering RNA.
Discussion
Osteosarcoma could induce bone damage and can lead to lung metastasis (26). The main features of osteosarcoma include the presence of mesenchymal spindle cells and production of the bone matrix (27). In terms of age distribution, patients with osteosarcoma are mainly adolescents and the elderly (28,29). The prognosis of patients with osteosarcoma has not been well improved or managed in the past 30 years (30). The genetic and biological complexity of osteosarcoma may be the main cause of this prognosis (31).
LncRNAs play critical roles during the development of multiple types of cancers (32,33). A study suggested that lower levels of lncRNA growth arrest-specific 5 promoted the proliferation and metastasis of ovarian cancer by targeting and suppressing the expression of miR-196-5p (34). In addition, lncRNA small nucleolar RNA host gene (SNHG) 3 could also modulate the proliferation, migration and invasion of ovarian cancer cells by regulating the energy metabolism of these cells (35). During osteosarcoma development, higher lncRNA SNHG4 expression could also promote the proliferation and metastasis of osteosarcoma cells by sponging miR-224-3p (31). LncRNA prostate cancer-associated transcript 6 (PCAT6) also enhanced the proliferation and colony formation ability of prostate cancer cells (11). The expression of lncRNA PCAT6 was also higher in the lung cancer tissues compared with adjacent normal tissues (25,36). The present study found that the expression of lncRNA PCAT6 in osteosarcoma cells was higher compared with hFOB1.19 cells. In addition, the proliferation, migration and invasion of osteosarcoma cells was promoted following lncRNA PCAT6 overexpression. Additionally, lncRNA PCAT6 overexpression also enhanced the proliferation of osteosarcoma cells in vivo.
MDM2/p53 signaling was reported to be involved in the proliferation and apoptosis of osteosarcoma cells (37,38). However, how MDM2/p53 signaling is regulated in osteosarcoma remains unclear. Drugs targeting MDM-p53 interaction, such as nutlin-3a and RG-7112, have been designed as new drugs against well-differentiated liposarcomas and osteosarcomas (39). Blocking of MDM-p53 interaction exerted efficacy by suppressing osteosarcoma cell growth (38,40). LncRNAs have been considered as potential therapeutic targets for osteosarcoma and regulate osteosarcoma progression (41–43). MDM2 is implicated in regulating long noncoding RNA maternally expressed 3, which affected MMP2 and MMP9 expression (44). Therefore, it was hypothesized that MDM2 could regulate MMP2 and MMP9 levels to affect migration and invasion ability of osteosarcoma cells. As shown in several studies, lncRNA plasmacytoma variant translocation 1 (PVT1) regulated MMP2 and MMP9 expression and affected radiosensitivity in non-small-cell lung cancer and cell migration abilities in a murine abdominal aortic aneurysm model (45,46).
Furthermore, P53 and P21 expression could inhibit the proliferation and metastasis of cancer cells. P53 is a cancer suppressor gene that was identified in 1979 (47) and is a crucial tumor suppressor gene (48). P53 expression is declined in tumor tissues (49). Several studies revealed that P53 suppressed the proliferation of cancer cells by regulating P21 expression and inducing cell cycle arrest (50,51). MDM2 expression was also associated with the development of multiple types of cancer (52,53). A study revealed that higher levels of MDM2 promoted the proliferation and metastasis of colorectal tumor cells (54). miRNA-379-5p could also inhibit the proliferation and metastasis of bladder cancer cells by suppressing MDM2 expression (53). A study indicated that MDM2 could aggravate the symptoms of cancer by suppressing P53 expression (55). The present study found that MDM2 expression increased while the levels of P53 and P21 decreased following lncRNA PCAT6 overexpression. However, the proliferation, migration and invasion of osteosarcoma cells were inhibited after MDM2 knockdown. P53 and P21 expression was also rescued following MDM2 knockdown. These results indicated that lncRNA PCAT6 promoted the proliferation, migration and invasion of osteosarcoma cells by promoting MDM2 expression and inhibiting P53 and P21 expression.
However, higher MDM2 levels were expressed in MG63 cells, which may be due to lncRNA PCAT6 upregulating MDM2 expression. Therefore, lncRNA PCAT6, which was reported to regulate miRNA levels, affected the expression of MDM2 possibly through miRNAs, (56–58). Additionally, in MG63 cells, MDM2 was regulated by proteins including Ras-ERK1/2 signaling or GRIM-19 (59,60). Thus, MDM2 could be regulated by lncRNA PCAT6 possibly attributing to the regulation of lncRNA PCAT6 on the upstream proteins of MDM2.
Additionally, the present study detected the effects of lncRNA PCAT6 on the proliferation and metastasis of osteosarcoma cells. The results indicated that lncRNA PCAT6 enhanced the proliferation, migration and invasion of osteosarcoma cells by promoting MDM2 expression and therefore inhibiting the expression of P53 and P21. This conclusion may provide a new target and therapy for the clinical treatment of osteosarcoma.
Acknowledgements
Not applicable.
Funding
This study was supported by the Science and Technology Project of Jiaxing (grant no. 2019AY32030), the Key Discipline of Jiaxing Oncology Medicine Construction Project (grant no. 2019-zc-11), Key Laboratory of Precision Treatment for Lung Cancer in Jiaxing, the Early Diagnosis and Comprehensive Treatment of Lung Cancer Innovation Team Building Project, Zhejiang North Regional Anaesthesia Special Disease Center and Clinical Research Project in Medical Committee of Zhejiang Province (grant no. 2013ZYC-A89).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XG, YX and JZ wrote the manuscript and performed the experiments. XG and JZ made substantial contributions to the conception and design of the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were performed in accordance with the animal experimental guidelines set by the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The study was approved by the Experimental Animal Ethical Committee of Zhejiang Hospital (approval no. JUMC2019-019).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ando K, Heymann MF, Stresing V, Mori K, Rédini F, Heymann D. Current therapeutic strategies and novel approaches in osteosarcoma. Cancers (Basel) 2013;5:591–616. doi: 10.3390/cancers5020591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: Current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 4.Kong G, Qi XJ, Wang JF. Effect of lncRNA LET on proliferation and invasion of osteosarcoma cells. Eur Rev Med Pharmacol Sci. 2018;22:1609–1614. doi: 10.26355/eurrev_201803_14567. [DOI] [PubMed] [Google Scholar]
- 5.Yan L, Wu X, Liu Y, Xian W. LncRNA Linc00511 promotes osteosarcoma cell proliferation and migration through sponging miR-765. J Cell Biochem. 2018 Dec 28; doi: 10.1002/jcb.27999. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 6.Li X, Wu Z, Fu X, Han W. Long noncoding RNAs: Insights from biological features and functions to diseases. Med Res Rev. 2013;33:517–553. doi: 10.1002/med.21254. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Shi D, Wu F, Mu S, Hu B, Zhong B, Gao F, Qing X, Liu J, Zhang Z, Shao Z. LncRNA AFAP1-AS1 promotes tumorigenesis and epithelial-mesenchymal transition of osteosarcoma through RhoC/ROCK1/p38MAPK/Twist1 signaling pathway. J Exp Clin Cancer Res. 2019;38:375. doi: 10.1186/s13046-019-1363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Y, Feng SJ, Qiu S, Shao N, Zheng JH. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2017;21:3176–3184. [PubMed] [Google Scholar]
- 10.Liang H, Yu T, Han Y, Jiang H, Wang C, You T, Zhao X, Shan H, Yang R, Yang L, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer. 2018;17:119. doi: 10.1186/s12943-018-0870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong FR, Lv YH, Yao HM, Zhang HY, Zhou Y, Liu SE. LncRNA PCAT6 promotes occurrence and development of ovarian cancer by inhibiting PTEN. Eur Rev Med Pharmacol Sci. 2019;23:8230–8238. doi: 10.26355/eurrev_201910_19132. [DOI] [PubMed] [Google Scholar]
- 13.Shi X, Liu Z, Liu Z, Feng X, Hua F, Hu X, Wang B, Lu K, Nie F. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177–187. doi: 10.1016/j.ebiom.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu C, Huang L, Xu F, Li P, Li P, Hu F. LncRNA PCAT6 promotes tumor progression in osteosarcoma via activation of TGF-β pathway by sponging miR-185-5p. Biochem Biophys Res Commun. 2020;521:463–470. doi: 10.1016/j.bbrc.2019.10.136. [DOI] [PubMed] [Google Scholar]
- 15.Han R, Huang G, Wang Y, Xu Y, Hu Y, Jiang W, Wang T, Xiao T, Zheng D. Increased gene expression noise in human cancers is correlated with low p53 and immune activities as well as late stage cancer. Oncotarget. 2016;7:72011–72020. doi: 10.18632/oncotarget.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X, Hu Y, Wu J, Shi L, Zhu L, Xi PW, Wei JF, Ding Q. RBMS2 inhibits the proliferation by stabilizing P21 mRNA in breast cancer. J Exp Clin Cancer Res. 2018;37:298. doi: 10.1186/s13046-018-0968-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou PF, Jiang T, Chen F, Shi PC, Li HQ, Bai J, Song J. KIF4A facilitates cell proliferation via induction of p21-mediated cell cycle progression and promotes metastasis in colorectal cancer. Cell Death Dis. 2018;9:477. doi: 10.1038/s41419-018-0550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim EM, Jung CH, Kim J, Hwang SG, Park JK, Um HD. The p53/p21 complex regulates cancer cell invasion and apoptosis by targeting Bcl-2 family proteins. Cancer Res. 2017;77:3092–3100. doi: 10.1158/0008-5472.CAN-16-2098. [DOI] [PubMed] [Google Scholar]
- 19.Seipel K, Marques MAT, Sidler C, Mueller BU, Pabst T. The cellular p53 inhibitor MDM2 and the growth factor receptor FLT3 as biomarkers for treatment responses to the MDM2-inhibitor idasanutlin and the MEK1 inhibitor cobimetinib in acute myeloid leukemia. Cancers (Basel) 2018;10:170. doi: 10.3390/cancers10060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Wang DD, Wu YP, Su D, Zhou TY, Gai RH, Fu YY, Zheng L, He QJ, Zhu H, Yang B. MDM2 promotes epithelial-mesenchymal transition and metastasis of ovarian cancer SKOV3 cells. Br J Cancer. 2017;117:1192–1201. doi: 10.1038/bjc.2017.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deben C, Deschoolmeester V, Lardon F, Rolfo C, Pauwels P. TP53 and MDM2 genetic alterations in non-small cell lung cancer: Evaluating their prognostic and predictive value. Crit Rev Oncol Hematol. 2016;99:63–73. doi: 10.1016/j.critrevonc.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Mizoshiri N, Shirai T, Terauchi R, Tsuchida S, Mori Y, Hayashi D, Kishida T, Arai Y, Mazda O, Nakanishi T, Kubo T. The tetraspanin CD81 mediates the growth and metastases of human osteosarcoma. Cell Oncol (Dordr) 2019;42:861–871. doi: 10.1007/s13402-019-00472-w. [DOI] [PubMed] [Google Scholar]
- 24.Benassi MS, Gamberi G, Magagnoli G, Molendini L, Ragazzini P, Merli M, Chiesa F, Balladelli A, Manfrini M, Bertoni F, et al. Metalloproteinase expression and prognosis in soft tissue sarcomas. Ann Oncol. 2001;12:75–80. doi: 10.1023/A:1011184413602. [DOI] [PubMed] [Google Scholar]
- 25.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Knockdown of long noncoding RNA PCAT6 inhibits proliferation and invasion in lung cancer cells. Oncol Res. 2016;24:161–170. doi: 10.3727/096504016X14618564639178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein MJ, Siegal GP. Osteosarcoma: Anatomic and histologic variants. Am J Clin Pathol. 2006;125:555–581. doi: 10.1309/UC6KQHLD9LV2KENN. [DOI] [PubMed] [Google Scholar]
- 27.Miller BJ, Gao Y, Duchman KR. Socioeconomic measures influence survival in osteosarcoma: An analysis of the national cancer data base. Cancer Epidemiol. 2017;49:112–117. doi: 10.1016/j.canep.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. 2009;125:229–234. doi: 10.1002/ijc.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger M, Fagioli F, Abate M, Riccardi R, Prete A, Cozza R, Bertulli R, Podda M, Ferrari S, Luksch R. Unusual sites of Ewing sarcoma (ES): A retrospective multicenter 30-year experience of the Italian association of pediatric hematology and oncology (AIEOP) and Italian sarcoma group (ISG) Eur J Cancer. 2013;49:3658–3665. doi: 10.1016/j.ejca.2013.06.045. [DOI] [PubMed] [Google Scholar]
- 31.Xu R, Feng F, Yu X, Liu Z, Lao L. LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and predicts poor survival and recurrence in human osteosarcoma. Cell Prolif. 2018;51:e12515. doi: 10.1111/cpr.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang L, Guo R, Yuan Z, Shi H, Zhang D. LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging miR-206 in ovarian cancer. Cell Physiol Biochem. 2018;49:1289–1303. doi: 10.1159/000493408. [DOI] [PubMed] [Google Scholar]
- 33.Wu W, Gao H, Li X, Zhu Y, Peng S, Yu J, Zhan G, Wang J, Liu N, Guo X. LncRNA TPT1-AS1 promotes tumorigenesis and metastasis in epithelial ovarian cancer by inducing TPT1 expression. Cancer Sci. 2019;110:1587–1598. doi: 10.1111/cas.14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H, Yu H, Zheng J, Ning N, Tang F, Yang Y, Wang Y. Lowly-expressed lncRNA GAS5 facilitates progression of ovarian cancer through targeting miR-196-5p and thereby regulating HOXA5. Gynecol Oncol. 2018;151:345–355. doi: 10.1016/j.ygyno.2018.08.032. [DOI] [PubMed] [Google Scholar]
- 35.Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150:343–354. doi: 10.1016/j.ygyno.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 36.Wan L, Zhang L, Fan K, Wang JJ. Diagnostic significance of circulating long noncoding RNA PCAT6 in patients with non-small cell lung cancer. Onco Targets Ther. 2017;10:5695–5702. doi: 10.2147/OTT.S149314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y, Lv C, Shi L, Tu G. MEG3 inhibits proliferation and invasion and promotes apoptosis of human osteosarcoma cells. Oncol Lett. 2018;15:1917–1923. doi: 10.3892/ol.2017.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang B, Fang L, Zhao H, Xiang T, Wang D. MDM2 inhibitor Nutlin-3a suppresses proliferation and promotes apoptosis in osteosarcoma cells. Acta Biochim Biophys Sin (Shanghai) 2012;44:685–691. doi: 10.1093/abbs/gms053. [DOI] [PubMed] [Google Scholar]
- 39.Wang S, Zhao Y, Aguilar A, Bernard D, Yang CY. Targeting the MDM2-p53 protein-protein interaction for new cancer therapy: Progress and challenges. Cold Spring Harb Perspect Med. 2017;7:a026245. doi: 10.1101/cshperspect.a026245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tovar C, Graves B, Packman K, Filipovic Z, Higgins B, Xia M, Tardell C, Garrido R, Lee E, Kolinsky K, et al. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013;73:2587–2597. doi: 10.1158/0008-5472.CAN-12-2807. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao W, Zhang D, Qin P, Zhang J, Cui X, Gao J, Wang J, Li J. Long non-coding RNA EPIC1 inhibits viability and invasion of osteosarcoma cells by promoting MEF2D ubiquitylation. Int J Biol Macromol. 2019;128:566–573. doi: 10.1016/j.ijbiomac.2019.01.156. [DOI] [PubMed] [Google Scholar]
- 43.Hu T, Fei Z, Su H, Xie R, Chen L. Polydatin inhibits proliferation and promotes apoptosis of doxorubicin-resistant osteosarcoma through LncRNA TUG1 mediated suppression of Akt signaling. Toxicol Appl Pharmacol. 2019;371:55–62. doi: 10.1016/j.taap.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Li Z, Yang L, Liu X, Nie Z, Luo J. Long noncoding RNA MEG3 inhibits proliferation of chronic myeloid leukemia cells by sponging microRNA21. Biomed Pharmacother. 2018;104:181–192. doi: 10.1016/j.biopha.2018.05.047. [DOI] [PubMed] [Google Scholar]
- 45.Wang D, Hu Y. Long non-coding RNA PVT1 competitively binds MicroRNA-424-5p to regulate CARM1 in radiosensitivity of non-small-cell lung cancer. Mol Ther Nucleic Acids. 2019;16:130–140. doi: 10.1016/j.omtn.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Zhang Z, Zou G, Chen X, Lu W, Liu J, Zhai S, Qiao G. Knockdown of lncRNA PVT1 inhibits vascular smooth muscle cell apoptosis and extracellular matrix disruption in a murine abdominal aortic aneurysm model. Mol Cells. 2019;42:218–227. doi: 10.14348/molcells.2018.0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 48.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gunia S, Kakies C, Erbersdobler A, Hakenberg OW, Koch S, May M. Expression of p53, p21 and cyclin D1 in penile cancer: p53 predicts poor prognosis. J Clin Pathol. 2012;65:232–236. doi: 10.1136/jclinpath-2011-200429. [DOI] [PubMed] [Google Scholar]
- 51.Li M, Li L, Zhang L, Hu W, Shen J, Xiao Z, Wu X, Chan FL, Cho CH. 1,25-Dihydroxyvitamin D3 suppresses gastric cancer cell growth through VDR- and mutant p53-mediated induction of p21. Life Sci. 2017;179:88–97. doi: 10.1016/j.lfs.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 52.Abolhasani M, Salarinejad S, Asgari M. P53 and MDM2 over-expression and five-year survival of kidney cancer patients undergoing radical nephrectomy-iranian experience. Asian Pac J Cancer Prev. 2015;16:5043–5047. doi: 10.7314/APJCP.2015.16.12.5043. [DOI] [PubMed] [Google Scholar]
- 53.Wu D, Niu X, Tao J, Li P, Lu Q, Xu A, Chen W, Wang Z. MicroRNA-379-5p plays a tumor-suppressive role in human bladder cancer growth and metastasis by directly targeting MDM2. Oncol Rep. 2017;37:3502–3508. doi: 10.3892/or.2017.5607. [DOI] [PubMed] [Google Scholar]
- 54.Chaar I, Amara S, Khiari M, Ounissi D, Dhraif M, Ben Hamida AE, Gharbi L, Mzabi S, Bouraoui S. Relationship between MDM2 and p53 alterations in colorectal cancer and their involvement and prognostic value in the Tunisian population. Appl Immunohistochem Mol Morphol. 2013;21:228–236. doi: 10.1097/PAI.0b013e31825f4e20. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, Wang X, Wang G, Yang Y, Yuan Y, Ouyang L. The past, present and future of potential small-molecule drugs targeting p53-MDM2/MDMX for cancer therapy. Eur J Med Chem. 2019;176:92–104. doi: 10.1016/j.ejmech.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 56.Cui LH, Xu HR, Yang W, Yu LJ. lncRNA PCAT6 promotes non-small cell lung cancer cell proliferation, migration and invasion through regulating miR-330-5p. Onco Targets Ther. 2018;11:7715–7724. doi: 10.2147/OTT.S178597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu H, Zou Q, He H, Liang Y, Lei M, Zhou Q, Fan D, Shen L. Long non-coding RNA PCAT6 targets miR-204 to modulate the chemoresistance of colorectal cancer cells to 5-fluorouracil-based treatment through HMGA2 signaling. Cancer Med. 2019;8:2484–2495. doi: 10.1002/cam4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xin Y, He X, Zhao W, Zhan M, Li Y, Xiao J, He K, Lu L. LncRNA PCAT6 increased cholangiocarcinoma cell proliferation and invasion via modulating miR-330-5p. Am J Transl Res. 2019;11:6185–6195. [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, Liu M, Liu W, Wang W. Ras-ERK1/2 signalling promotes the development of osteosarcoma through regulation of H4K12ac through HAT1. Artif Cells Nanomed Biotechnol. 2019;47:1207–1215. doi: 10.1080/21691401.2019.1593857. [DOI] [PubMed] [Google Scholar]
- 60.Chen W, Liu Q, Fu B, Liu K, Jiang W. Overexpression of GRIM-19 accelerates radiation-induced osteosarcoma cells apoptosis by p53 stabilization. Life Sci. 2018;208:232–238. doi: 10.1016/j.lfs.2018.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.