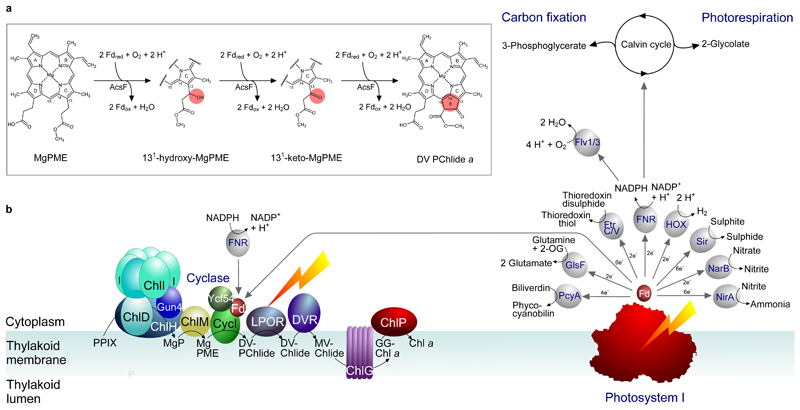
Fig. 6. Diagram depicting the Fd-dependent cyclase reaction catalysed by AcsF and the supply of reduced Fd, directly or indirectly, by PSI.
a, The updated sequence of cyclase reactions catalysed by AcsF, with the chemical change of the porphyrin substrate at each step highlighted in the pink circle. Fdred and Fdox represent reduced and oxidised Fd, respectively. b, The chlorophyll biosynthesis pathway is shown on the left, progressing from protoporphyrin IX (PPIX) to chlorophyll a (Chl a), via magnesium protoporphyrin IX (MgP), Mg-protoporphyrin IX monomethyl ester (MgPME), 3,8-divinyl protochlorophyllide a (DV-PChlide), divinyl chlorophyllide a (DV-Chlide), monovinyl chlorophyllide a (MV-Chlide), and geranylgeranyl-chlorophyll a (GG-Chl a). ChlH, D, I are subunits, and Gun4 is an accessory protein, of the magnesium chelatase complex; ChlM is the MgP methyltransferase; CycI is the counterpart of the AcsF cyclase in cyanobacteria and plants, shown here with the accessory protein Ycf54 and Fd; LPOR is the light-dependent PChlide oxidoreductase; DVR is the divinyl reductase, the cyanobacterial version (BciB) of which requires Fd, whereas the plant DVR does not. ChlG is the chlorophyll synthase and ChlP is the geranylgeranyl reductase. The diagram (right) depicts a possible direct link between PSI and chlorophyll biosynthesis, showing that PSI could provide reduced Fd for the cyclase reaction; FNR-based reduction of Fd is also depicted, corresponding to the in vitro assays in Fig. 3 and Extended Data Fig. 2 respectively. Reduced Fd also provides electrons for a variety of cellular functions, shown here for cyanobacterial metabolism and adapted from the diagram in ref. 41. PcyA, phycocyanobilin:Fd oxidoreductase; GlsF, Fd-dependent glutamate synthase; FtrC/V, Fd:thioredoxin reductase; FNR, Fd:NADP+ reductase; Sir, Fd:sulfite reductase; NarB, nitrate reductase; NirA, nitrite reductase; Flv1/3, Flavodiiron 1/3; HOX, bi-directional hydrogenase.