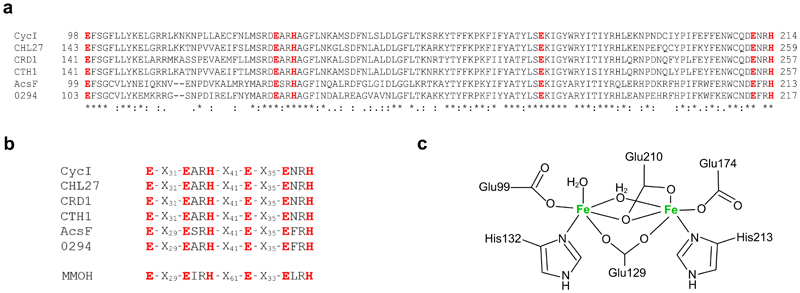
Extended Data Fig. 3. The diiron binding motif and proposed diiron ligation of AcsF.
a, Sequence alignments showing the conserved diiron binding motif of AcsF proteins. Sequences are from Synechocystis sp. PCC 6803 (CycI, BAA16583), Arabidopsis thaliana (CHL27, NP_191253), Chlamydomonas reinhardtii (CRD1, XP_001692557; CTH1, XP_001691047), Rubrivivax gelatinosus IL144 (AcsF, BAL96694) and Rhodobacter sphaeroides 2.4.1 (0294, abbreviated for RSP_0294, YP_353369). Conserved, highly similar and similar residues are marked with asterisks, colons and full stops, respectively. The putative diiron ligands are in red and bold. Full-length protein sequences were used for alignments but for clarity, only the putative diiron binding motifs with the residue range indicated, are shown. b, Sequence homologies between the diiron binding motifs of AcsF proteins and the soluble methane monooxygenase hydroxylase subunit from Methylococcus capsulatus Bath (MMOH, P22869). c, Proposed coordination of the diiron ligands of AcsF at the diferrous state based on the crystal structure of MMOH (PDB, 1FYZ)51.