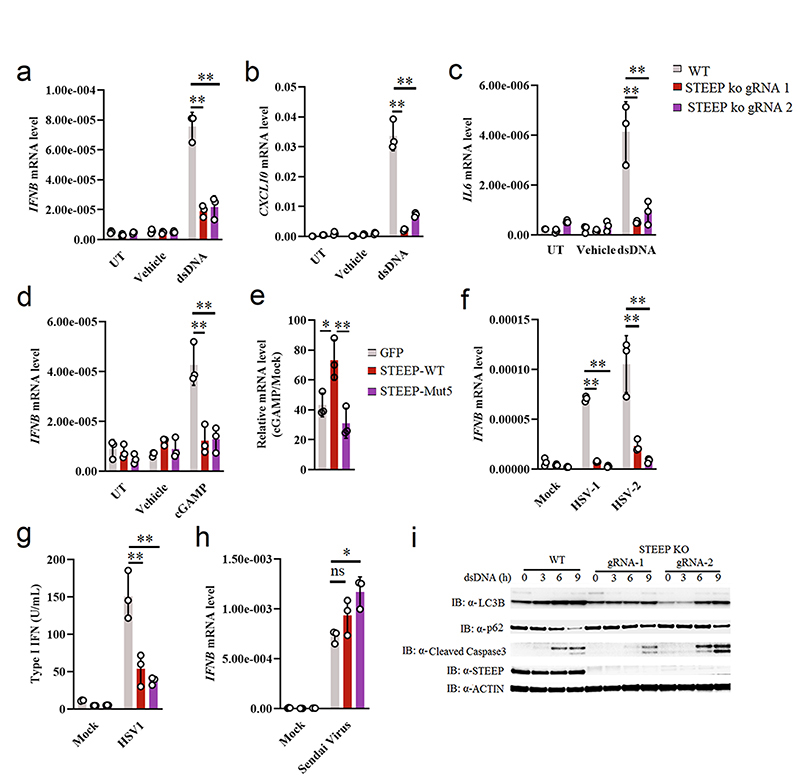
Figure 2. STEEP is essential for IFN induction through the cGAS-STING pathway.
(a-d) WT and STEEP-deficient THP-1 cells were treated with (a-c) dsDNA (500 ng/ml, 6h) (**P < 0.01), (d) 2’3’ cGAMP (100 nM, 2h) (**P = 0.0022, **P = 0.0025, left to right). Total RNA was isolated, and IFNB, CXCL10, and IL6 mRNA were measured as indicated by RT-qPCR. (e) STEEP-deficient THP-1 cells transfected with mRNA encoding GFP, STEEP-WT or STEEP-Mut5, and subsequently stimulated with cGAMP (100 nM, 2h) (**P = 0.032, **P = 0.0068, left to right). IFNB was measured by RT-qPCR. (f-h) WT and STEEP-deficient THP-1 cells were infected with (f-g) HSV1 or HSV2 (both MOI 1, 12 h) (**P < 0.01), or (h) Sendai virus (MOI 1, 12 h) (ns P = 0.19, *P = 0.014, left to right). . Total RNA or culture supernatants were isolated, and IFNB mRNA and type I IFN bioactivity were measured as indicated by RT-qPCR and biological assay, respectively. (n = 3, biologically independent samples) (i) WT and STEEP-deficient THP-1 cells were treated with dsDNA (2 ug/ml) for the indicated time intervals, and levels of LC3, p62, cleaved caspase 3 (CC3), STEEP and actin were determined by immunoblotting. Representative blot from three independent experiments with similar results are shown. Data in panel a-h are shown as means of biological sample triplicates +/- st.dev. (n = 3 biologically independent samples for all data sets in the figure). Statistical analysis of data presented in panel a-h was performed using two-tailed one-way ANOVA test.