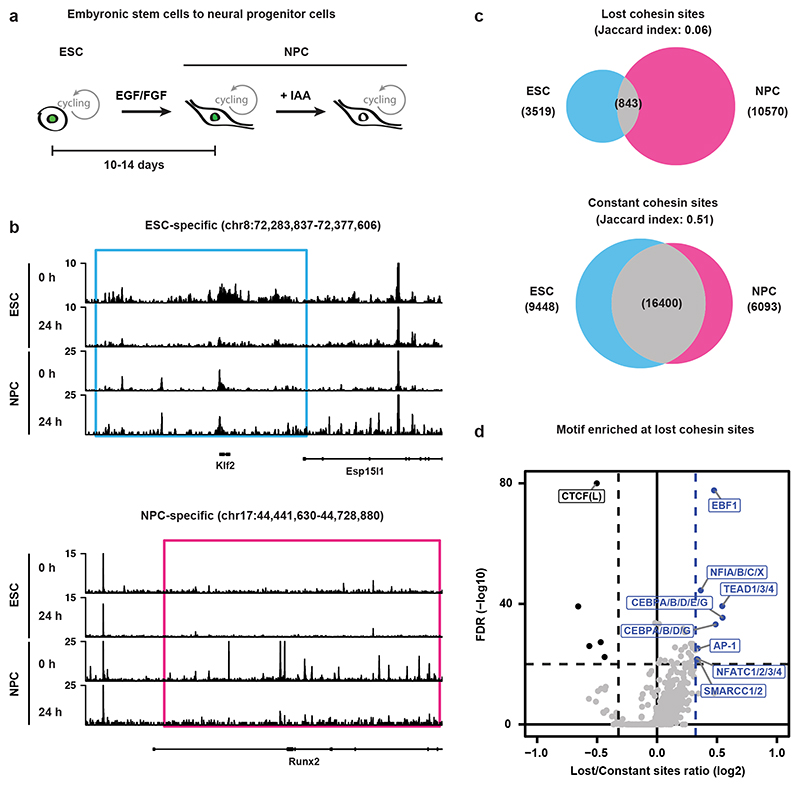
Fig. 5. Cohesin binding redistribution is a feature in differentiated cells.
a, WAPL-AID cells were differentiated to neural precursor cells using a standard differentiation protocol37. b, Example regions show the ChIP-seq tracks of RAD21 for treated and untreated WAPL-AID mESCs and NPCs. Light blue and pink rectangles highlight RAD21 lost regions in mESCs and NPCs, respectively. c, Venn diagram showing overlap between lost and constant cohesin binding sites in mESCs and NPCs. Lost cohesin sites are regions bound in the presence of WAPL, but not bound in the absence of WAPL. Constant cohesin sites are regions bound in the presence and absence of WAPL. Peak calling was performed on a subset of 7 M sequencing reads for all the samples. d, Motif enrichment analysis for lost cohesin binding sites. Sites bound preferentially in untreated cells (“Lost”) and sites bound stably in both treated and untreated cells (“Constant”) were analyzed with GimmeMotifs63. P values are calculated using the two-sided Fisher exact test for motif frequency for lost and constant peaks for every motif. Log2 fold-change of motif frequency is determined by calculating the ratio of relative motif frequency (i.e. corrected for the total motif frequency of all motifs) between lost and constant peaks (see Methods for details on fold-change and Pvalue calculation).