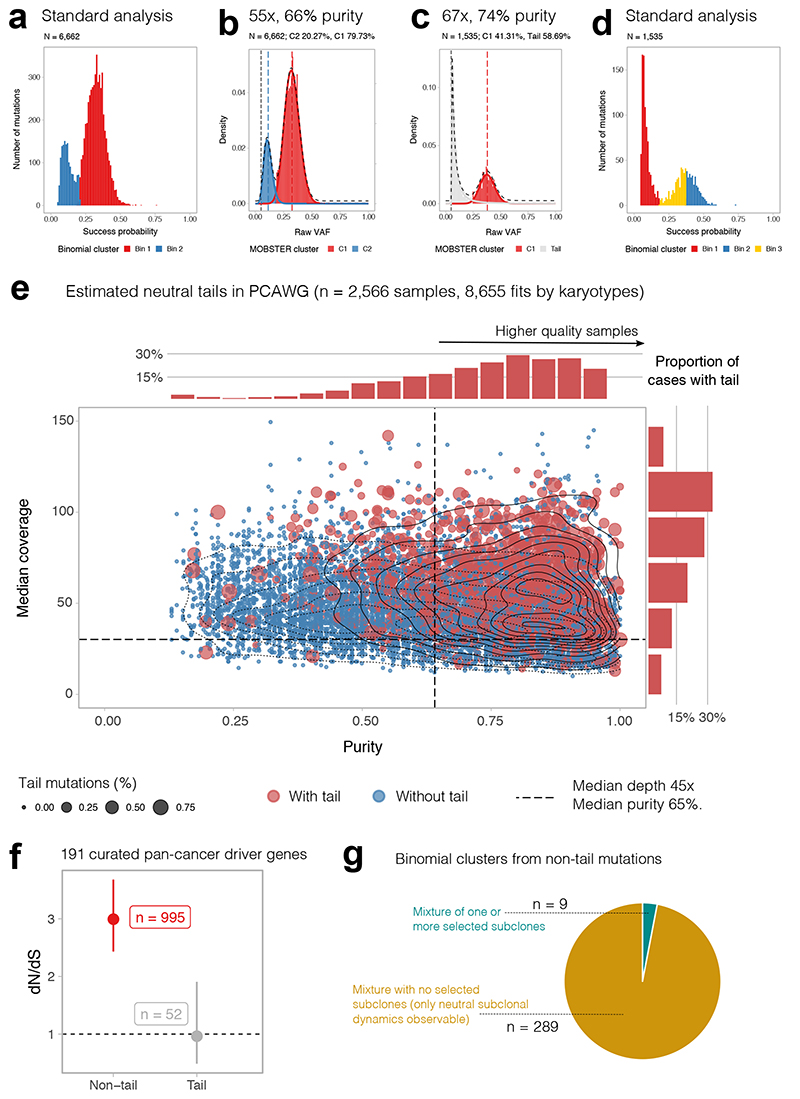
Figure 4. Analysis of 2,566 whole-genomes from PCAWG with MOBSTER.
(a) Fit of a PCAWG25 tumor with 55x coverage and 66% purity using standard methods. (b) At this data resolution, neutral tails are under-sampled (Figure 1j,k) and cannot be distinguished from selected subclones. (c) In PCAWG cases with higher coverage (67x) and purity (74%), neutral tails can be clearly detected using MOBSTER. (d) Analysis of the same tumor with standard methods would have identified multiple subclonal clusters, including a cluster of neutral tail mutations. (e) We analyzed n=2,566 PCAWG samples, plotted here for purity vs coverage. Blue dots are tumors where MOBSTER cannot fit a tail. Red cases have a neutral tail. Percentage of tail mutations determines dot size. The marginal histograms report the normalized number of cases with tail. (f) We focused on the 902 diploid cases with coverage > 30x and purity > 65% (median of the cohort) where we could fit a tail. Using a panel of 191 pan-cancer driver genes, we show that tail mutations have dN/dS~1, providing no evidence of positive selection (point estimate and Confidence Intervals from dndscv). Clonal and subclonal non-tail mutations show dN/dS> 1, consistent with positive selection. (g) If we take the 298 diploid cases with a tail containing at least 10% of the total mutational burden, we find evidence of a selected subclone only in 9 cases (3% of tumors). Similar proportions are obtained if we impose a 5% or 2% cutoff on the size of the tail. See Supplementary Figures 29-31.