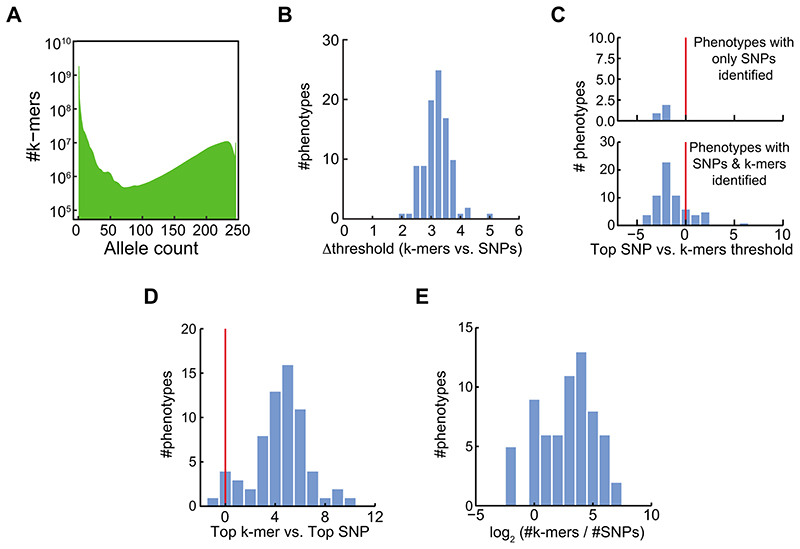
Extended Data Fig. 9. Comparison of SNP- and k-mer-based GWAS in tomato.
(A) Histogram of k-mers allele counts for tomato accessions.
(B) Histogram of difference between threshold values of SNPs and k-mers for tomato phenotypes.
(C) Histogram of the top SNP p-value divided by the k-mers defined threshold, in -log10, for tomato phenotypes. Plotted for phenotypes with only identified SNPs (upper panel) or for phenotypes with both SNPs and k-mers identified (lower panel).
(D) Histogram of the difference between top (-log10) p-values in the two methods for tomato phenotypes.
(E) Histogram of the number of identified k-mers vs. identified SNPs for tomato phenotypes.