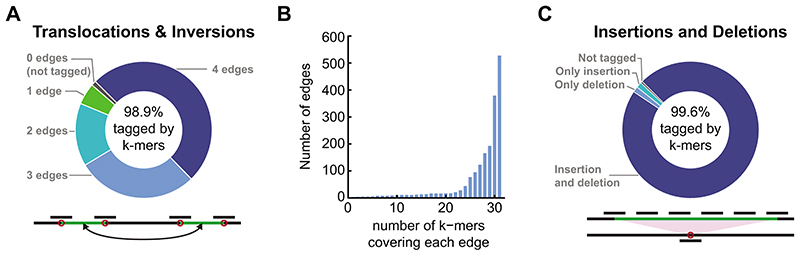
Extended Data Fig. 2. Genome-wide evaluation of k-mer potential to detect SVs in well-characterized genomes.
(A) For every translocation or inversion, previously identified22 between the Col-0 reference genome or the Ler genome we evaluate if it is tagged by 31 bp k-mers. Each translocation or inversion will affect 4 edges between the translocated fragment and the neighbouring genomic regions (bottom panel). For every previously identified translocation or inversion, the number of edges (0-4) which are tagged by k-mers unique to one genome were counted. Only 1.1% of these SVs were not tagged by any k-mer unique to one genome (upper panel).
(B) For every edge tagged by k-mers, described in A, we plot the number of k-mers unique to one genome which tagged it. The histogram is enriched with edges covered by the maximal number of k-mers, 31.
(C) Evaluating the potential to tag by k-mers long insertions/ deletions between the well characterized genomes of Col-0 and Ler22. While in the genome with the apparent deletion only the junction between the two fragments will be tagged by unique k-mers, in the genome with the apparent insertion, the entire insert will be tagged (bottom panel). Only 0.4% of the previously characterized long insertions/deletions are not tagged by unique k-mers.