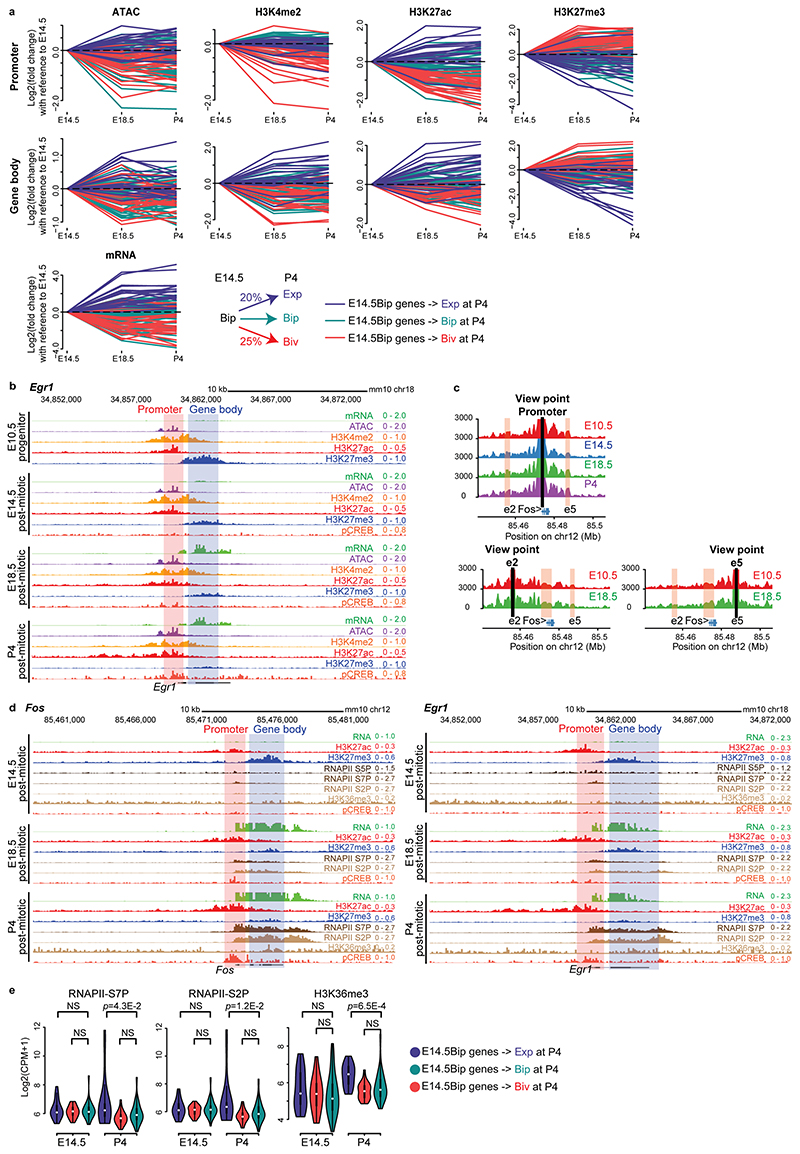
Extended Data Fig. 6. Developmental dynamics of the bipartite chromatin signature.
a, Developmental dynamics of chromatin profiles (ATAC-seq and ChIP-seq signals in promoter and gene body regions) and mRNA levels of E14.5Bip genes in E14.5, E18.5 and P4 Drg11vPrV-ZsGreen/+ barrelette neurons. Log2 fold changes are calculated with reference to E14.5. At P4, 20% of E14.5Bip genes become expressed (Exp, i.e. RPKM >= 3 at P4), 25% become bivalent (Biv, red dots in Fig. 3d), and the rest (55%) remain bipartite (Bip), (blue, red and green lines, respectively). Bottom: summary diagram. b, Genome browser view of the Egr1 locus at the E10.5, E14.5, E18.5 and P4 stages. c, 3D interaction map (4C-seq) using the Fos promoter (top left), enhancer 2 (e2, bottom left) and enhancer 5 (e5, bottom right) as viewpoints in E10.5, E14.5, E18.5 and P4 hindbrain tissue. Normalized read per 4C fragment is visualized. d, Genome browser views of Fos and Egr1 at the E14.5, E18.5 and P4 stages. e, Violin plots showing transcription end site (TES, Methods) RNAPII-S7P (left), RNAPII-S2P (middle), and H3K36me3 (right) levels of E14.5Bip genes at E14.5 and P4 (see a). E14.5Bip genes that become expressed (Exp, blue, n = 25) at P4 displayed significantly higher levels of RNAPII-S7P, -S2P and H3K36me3 marks as compared to E14.5Bip genes that become bivalent (Biv, red, n = 20) or remain bipartite (Bip, green, n = 55) at P4: violin deviations between groups are to be compared within the same time point since the time points reflect different batches. Plots extend from the data minima to the maxima with the white dot indicating median, the box showing the interquartile range and whiskers extending to the most extreme data point within 1.5X the interquartile range. P values are from two-sided Wilcoxon’s tests. NS: not significant (P > 0.05).