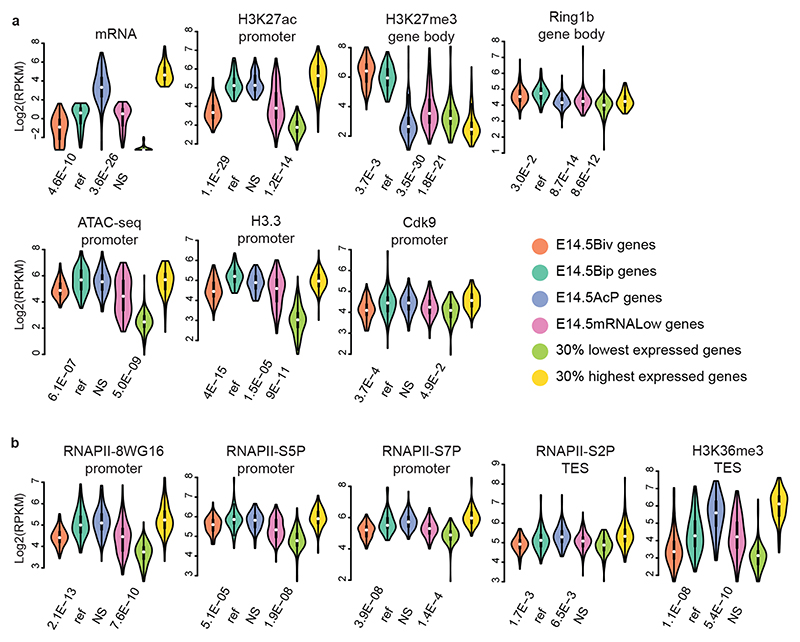
Fig. 4. RNA PolII transcripts of bipartite genes are not efficiently processed to productive mRNA.
a and b, Violin plots of RNA expression (RNA-seq) and chromatin features (ATAC-seq and ChIP-seq) of bivalent, bipartite, and control gene sets, as indicated. Comparison of Drg11vPrV-ZsGreen/+ barrelette neuron E14.5 bivalent genes (E14.5Biv, orange), E14.5 bipartite genes (E14.5Bip, green), E14.5 non-Bip genes with Bip-matching promoter H3K27ac levels (E14.5AcP, blue), E14.5 non-Bip genes with E14.5Bip-matching mRNA levels (E14.5mRNALow, magenta), 30% lowest (light green) and 30% highest (yellow) expressed genes illustrating the maximal signal range (Methods). Each gene set contains n = 97 genes (Methods). Plots extend from the data minima to the maxima with the white dot indicating median, the box showing the interquartile range and whiskers extending to the most extreme data point within 1.5× the interquartile range from the box. P values are from a two-sided Wilcoxon’s tests between each gene set and the E14.5Bip gene set, labeled as ‘ref’ for reference. NS: not significant (P > 0.05).