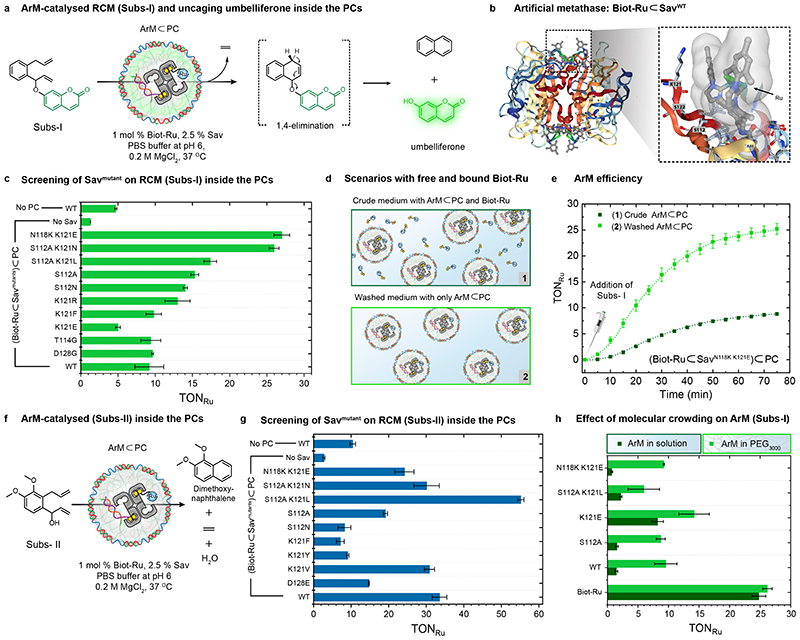
Fig. 3. Intraprotocellular ring-closing metathesis (RCM) of Subs-I and Subs-II catalysed by genetically-engineered ArMs.
(a) Schematic representation of the RCM-triggered uncaging of umbelliferone inside the PCs from a caged precursor (Subs-I) (b) X-ray crystal structure (PDB: 5IRA) of the artificial metathase Biot-Ru⊂SavWT. Biot-Ru and proximal protein residues are depicted as ball-and-stick and stick, respectively. (c) Genetic optimization of ArM for the uncaging of umbelliferone from Subs-I. The TONRu are based on yields of umbelliferone determined by fluorescence spectroscopy. Data are the means and standard deviation of duplicate reactions. (d) Schematic representation of two experiments: (1) RCM is performed using a crude mixture of free Biot-Ru (1 mol% catalyst loading with respect to the [Subs-I]) and ArM⊂PC, (2) RCM is performed using ArM⊂PC after a washing step to remove unbound Biot-Ru (i.e., centrifugation followed by re-dispersion). The Ru content of these samples was determined by ICP-MS (Supplementary Table 2). (e) Comparison of the turnover number (TONRu) of ArM⊂PC vs. free Biot-Ru, revealing the effect of the environment resulting from compartmentalization within Sav⊂PCs. (f) Schematic representation of the ArM-catalysed RCM of a dimethoxynaphthalene precursor (Subs-II) inside PCs. (g) Genetic optimization of the ArM for the RCM of Subs-II inside PCs. The TONRu are based on yields of dimethoxynaphthalene determined by GC-MS using an internal standard. Data are the means and standard deviation of duplicate reactions. (h) Crowding effect on the TONRu resulting from addition of 50 mg/mL PEG (3000 Da) for different mutants, the WT and the free co-factor Biot-Ru.