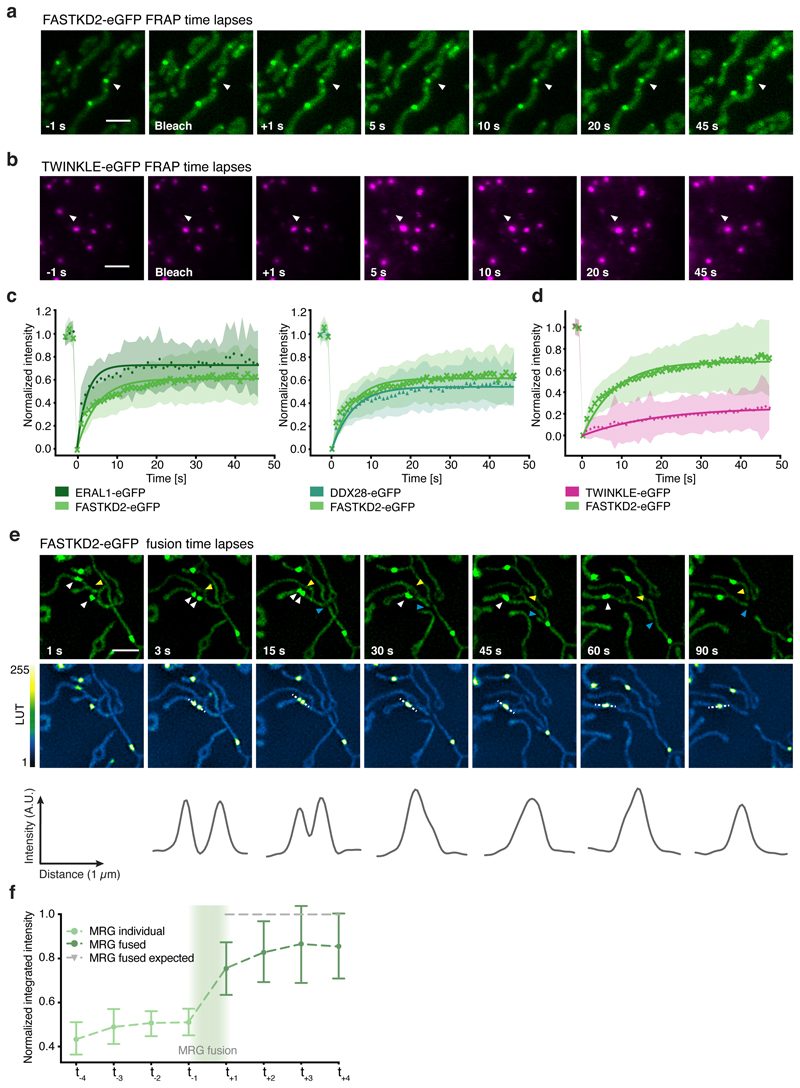
Fig. 2. MRGs exchange content and undergo fusion in live COS-7 cells.
a, b, Examples of FRAP time lapse series of MRGs (stable expression, FASTKD2-eGFP, green) or nucleoids (transient transfection, TWINKLE-eGFP, magenta), from datasets represented in (c) and (d), respectively. An individual MRG or nucleoid (arrowhead) was partially photobleached to allow tracking during recovery. Scale bars: 2 μm. c, d, FRAP intensity where symbols represent mean data points, lines are single exponential fits, and shaded areas standard deviations each time point. FASTKD2- (n = 44 MRGs examined over 8 independent experiments), ERAL1- (n = 17 MRGs examined over 3 independent experiments), DDX28- (n = 17 MRGs examined over 3 independent experiments) (c) and TWINKLE-eGFP (n = 50 nucleoids examined over 5 independent experiments) (d). e, Representative SIM time lapse series of an MRG fusion event (white arrowheads) in cells stably expressing FASTKD2-eGFP. Cells were imaged at 1/3 Hz. Yellow and blue arrowheads highlight mitochondrial network dynamics (top). Line profiles of MRGs along the mitochondrial axis (middle, dashed lines) show the intensity values at each time point (bottom). Scale bar: 2 μm. f, Temporal evolution of the integrated intensity of MRGs in COS-7 before and after fusion. Pre-fusion integrated intensities were summed for each granule pair (n=9 biologically independent cells), and their sum used for normalisation (grey dashed line). Mean and SD for each MRG type are shown for four time-points before (light green, t-4 – t-1) as well as after fusion (dark green, t+1 – t+4). Statistical source data are provided in Source data fig. 2.