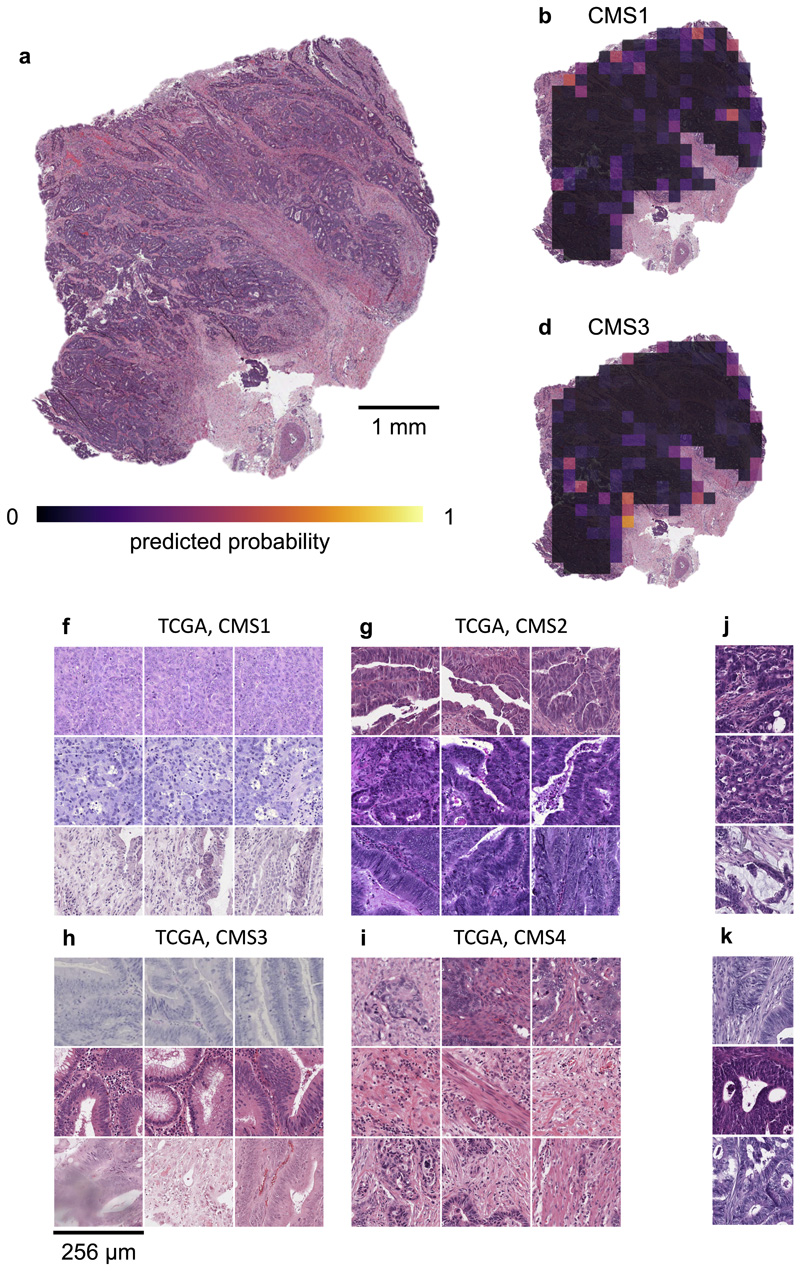
Fig. 5. Explainability of deep learning-based analysis of histological images.
Deep learning-based predictions were visualized through genotype maps and comparison of highly ranked image tiles. (a-e) Prediction maps for consensus molecular subtype (CMS) in colorectal cancer show spatially resolved prediction scores, unveiling intratumor heterogeneity of predicted genotype. As a generic tool, this visualization approach allows to identify spatial regions associated with a molecular feature. In this patient, the correct prediction of CMS4 correctly show that deep learning robustly predicts CMS from histology alone while highlighting potential intratumor heterogeneity (f-i) For each of the CMS classes, the most highly scored test set tiles are shown, enabling correlation of deep learning-predictions with histopathological features at high resolution. In this case, highly predicted CMS1 tiles contain numerous tumorinfiltrating lymphocytes while predicted CMS4 tiles contain abundant stroma, consistent with previous studies. (j-k) Highly scored tiles in the external test cohort DACHS for prediction of BRAF mutant and wild type (l-m) and CpG-island methylator phenotype (CIMP) high or non-CIMP.