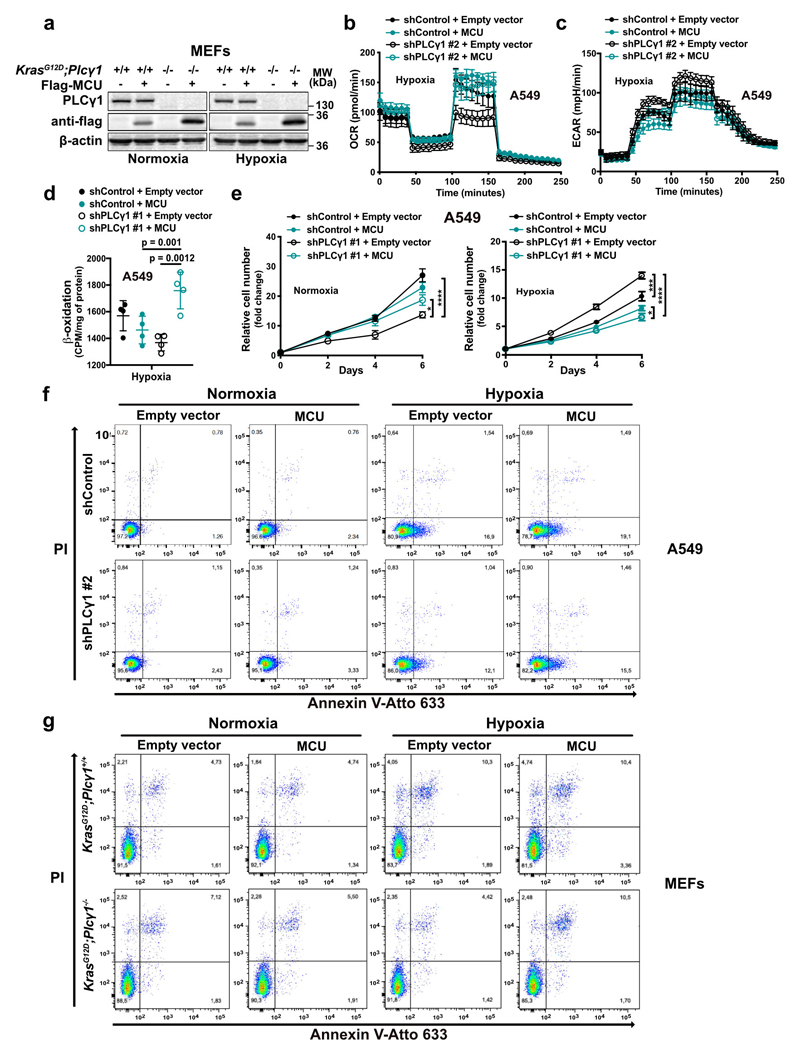
Extended Data Fig. 7. PLCγ1 suppression enhances cell proliferation and decreases cell death in hypoxia through impairment of Ca2+ entry into the mitochondria.
(a) Immunoblot analysis of the indicated targets in MEFs generated from a LSL-KrasG12D/WT;p53flox/flox;Plcγ1wt/wt and LSL-KrasG12D/WT;p53flox/flox;Plcγ1flox/flox mouse model, stably transduced with Cre recombinase (pMSCV-hygro-Cre) and transfected with either pcDNA3.1 empty vector or pcDNA3.1- MCU-flag plasmid and moved to hypoxia (1% O2) for additional 48h.
(b, c) Oxygen consumption rate (b) and extracellular acidification rate (c) of A549 cells transduced with either an empty vector (Tet-pLKO-puro, shControl) or a doxycycline-inducible shRNA against PLCγ1, treated with doxycycline for 24h, transfected with either pcDNA3.1 empty vector or pcDNA3.1-MCU-flag plasmid and incubated for 48h in hypoxia (3% O2). OCR was determined during sequential treatments (indicated with arrows) with oligomycin, FCCP and rotenone/antimycin (AA) at the indicated time. ECAR was determined during sequential treatments (indicated with arrows) with glucose (Glc), oligomycin and 2 deoxyglucose (2DG) at the indicated time; n = 5.
(d) Fatty acid β-oxidation of A549 cells transduced and treated as in (b); n = 4.
(e) Relative cell number of A549 cells transduced and treated as in (b); n = 3. Normoxia: * p = 0.03, hypoxia: * p = 0.014, *** p = 0.0005.
(f) Representative Annexin V-Atto 633/Propidium iodide (PI) flow cytometry analysis panels of A549 cells from experiment reported in main Fig. 5e; n = 3.
(g) Representative Annexin V-Atto 633/Propidium iodide (PI) flow cytometry analysis panels of MEF cells from experiment reported in main Fig. 5f; n = 3.
Graphical data are mean ± SD. Statistical analyses were done using one-way ANOVA; n, number of biologically independent samples. **** p < 0.0001. Statistical source data and unprocessed immunoblots are provided in Source Data Extended Data Fig. 7.