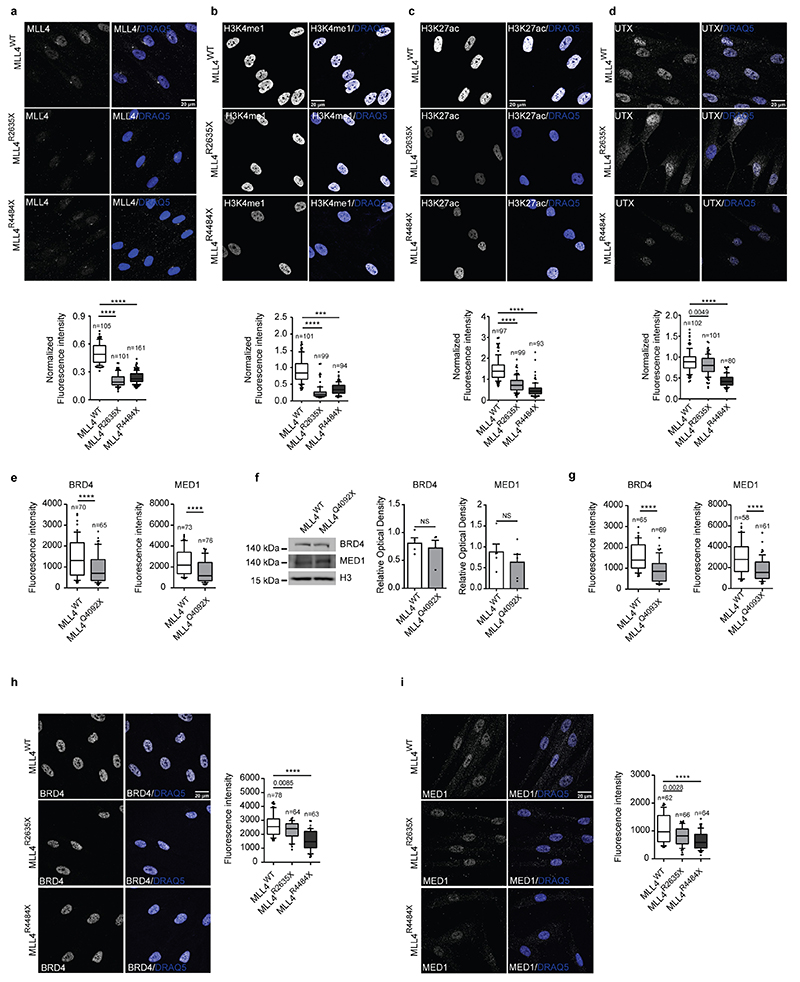
Extended Data Fig. 2.
a-d Representative images and quantifications of immunostaining for MLL4 (a), H3K4me1 (b), H3K27ac (c) and UTX (d) in primary fibroblasts from healthy donor or Kabuki patients. Scale bar, 20 μm. Box plots indicate the median (middle line), the first and third quartiles (box), and the 10th and 90th percentile (error bars) of the fluorescence intensity. The number of analyzed nuclei is reported in figure as n; unpaired two-tailed Student’s t-test was applied for statistical analysis (****P<0.0001).
e Quantifications of immunostaining for BRD4 and MED1 in WT and MLL4Q4092X MSCs grown on the same coverslips. WT MSCs were pre-labelled with CellTrace Violet Scale bar, 20 μm. Box plots indicate the median (middle line), the first and third quartiles (box), and the 10th and 90th percentile (error bars) of the fluorescence intensity. The number of analyzed nuclei is reported in figure as n; unpaired two-tailed Student’s t-test was applied for statistical analysis.
f Western Blot analysis of BRD4 and MED1 in WT and MLL4Q4092X MSCs; histone H3 was used as loading control. Signal quantifications are reported as bar plots. Data are means + SEM of 4 independent experiments for BRD4 and 5 independent experiments for MED1; one-tailed Student’s t-test was applied for statistical analysis.
g-i Representative images and quantifications of cluster intensity for BRD4 and MED1 immunostaining in WT and MLL4P4093X MSCs (g) on in primary fibroblasts from healthy donor or Kabuki patients (h-i). Box plots indicate the median (middle line), the first and third quartiles (box), and the 10th and 90th percentile (error bars) of the fluorescence intensity. The number of analyzed nuclei is reported in figure as n; unpaired two-tailed Student’s t-test was applied for statistical analysis (****P<0.0001).