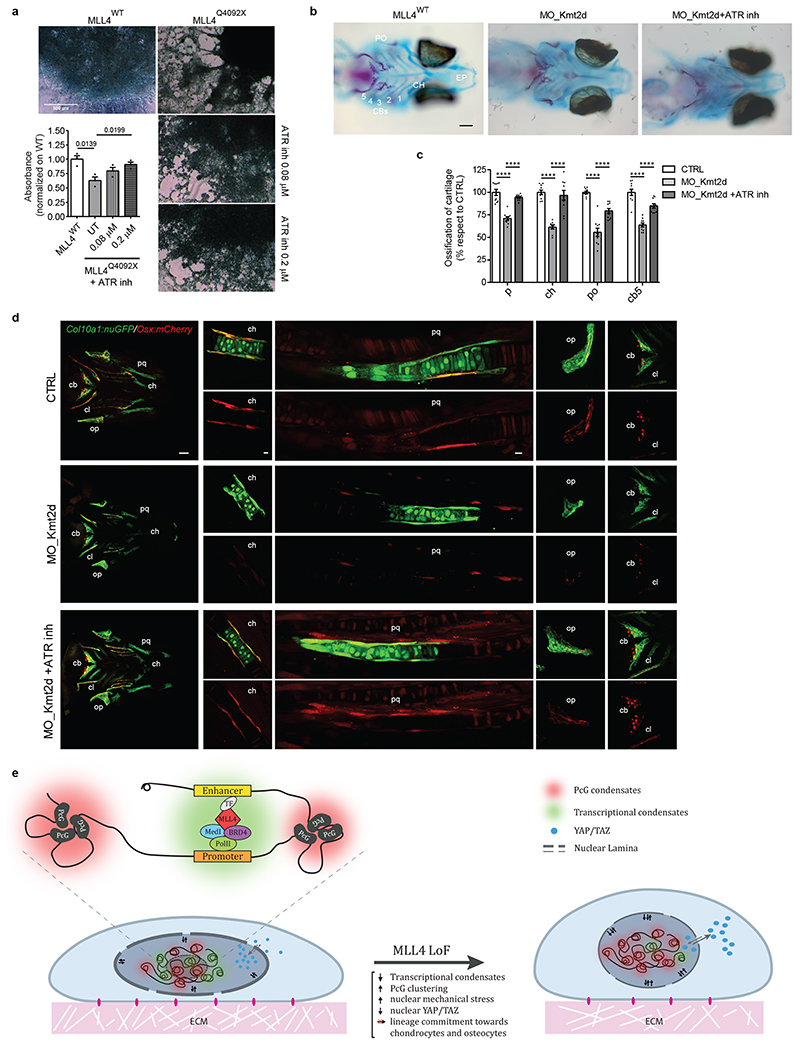
Fig. 8. MLL4-dependent mechanical properties in cell lineage commitment.
a Representative image of WT and MLL4Q4092X MSCs untreated or treated with different concentrations of ATR inhibitor (VE-922), after differentiation towards chondrocytes. Scale bar, 500 μm. Deposition of extracellular matrix and organization into a 3D network was detected by Alcian blue staining and quantified. Data are means + SEM (n = 3 independent experiments); unpaired two-tailed Student’s t-test was applied for statistical analysis.
b Alcian blue (cartilage) and alizarin Red (bone) staining of Ctrl, KMT2D 5’UTR-morpholino and VE822-treated KMT2D 5’UTR-morpholino medaka fish. Scale bar, 100μm.
c Bar plot of the relative quantification of Parasphenoid (P), Ceratohyal (CH), Paired Prootics (PO), Ceretobranchial 5 (CB5) mineralization in Ctrl, KMT2D 5’UTR-morpholino and KMT2D 5’UTR-morpholino-treated with ATR inhibitor. Values were expressed as percentage relative to the control. Data are means ± SEM (n = 10 biologically independent animals); Student’s t-test was applied for statistical analysis (**** P < 0.0001).
d Distribution of Col10a1:nuGFP- and Osx:mCherry-expressing premature osteoblasts in Ctrl (upper panels), KMT2D 5’UTR-morpholino (central panels) and VE822-treated KMT2D 5’UTR-morpholino medaka fish (lower panels). Scale bar: 100 μm.
e Schematic representation of MSCs alterations in response to MLL4 LoF. The upper panel represents the postulated interplay between transcription-associated and PcG condensates. The 3D chromatin compartmentalization, the intra-nuclear forces and the YAP/TAZ shuttling are shown in the representative cartoon.