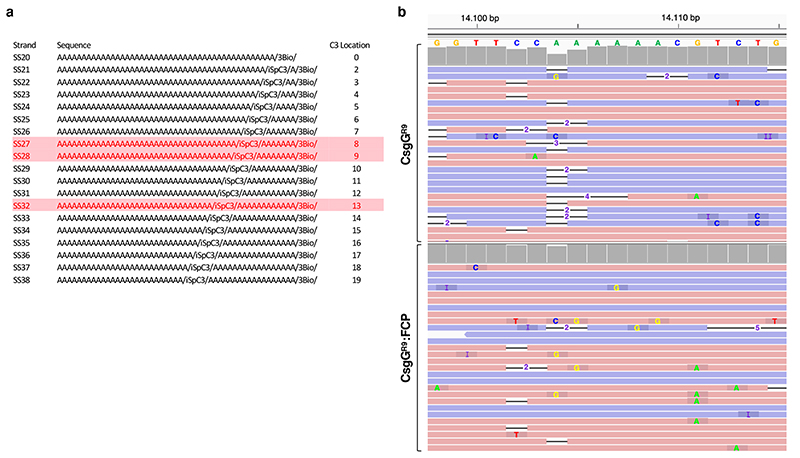
Extended Data Fig. 6. Constriction mapping oligos and single read basecalls for CsgGR9 and CsgGR9:FCP nanopores.
(a) Set of static polyA ssDNA oligonucleotides in which one base is missing from the DNA backbone (iSpc3). These oligos that have differing location of the abasic nucleotide, dubbed SS20 to SS38, were used to map the constriction position in CsgGF56Q or CsgGF56Q:FCP (Figure 3d). Biotin modification at the 3’ end of each strand is complexed with monovalent streptavidin to block translocation of the oligo and give a defined distance marker between the pore entrance (block site) and pore constriction (site of increased conductance when occupied by the abasic nucleotide; Figure 3c). SS27-SS28 and SS32 (highlighted red) have their abasic nucleotide located at the CsgG and FCP constriction, respectively (Figure 3d, e). (b) Comparison of errors in single read (n=26) basecalls from CsgGR9 and CsgGR9:FCP pores that have been aligned to a representative region of the E. coli reference genome sequence. The region displayed corresponds to the locus 14,098 to 14,115. The figure is plotted using the Integrative Genomics Viewer software33. Pink/purple bars correspond to single reads in the forward and reverse directions respectively. Black horizontal bars correspond to deletions in the basecalls, where the number corresponds to the number of deletions at the specific loci. Individual substitutions are labeled with the miscalled nucleotide (C in blue, T in red, G in orange and A in green). Insertions are labelled “I” (purple). Grey bars on top of the list of single reads of the CsgGR9 and CsgGR9:FCP pores correspond to the consensus accuracy per position.