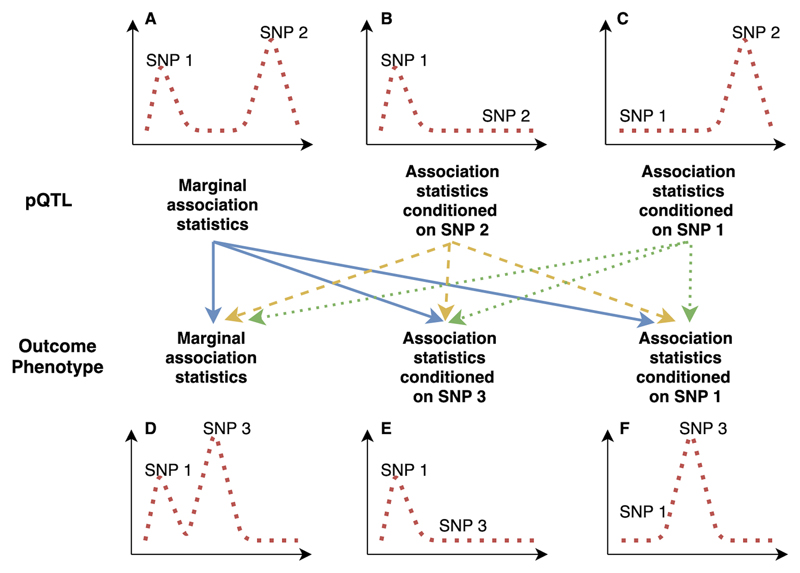
Figure 2. A demonstration of pairwise conditional and colocalization (PWCoCo) analysis.
Assume there are two conditional independent association pQTL signals (SNP 1 and SNP 2) and two conditional independent outcome signals (SNP 1 and SNP3) in the tested region. A naïve colocalization analysis using marginal association statistics will return weak evidence of colocalization (showed in regional plots A and D). By conducting the analyses conditioning on SNP 2 (plot B) and 1 (plot C) for the pQTLs and conditioning on SNP 1 (plot E) and 3 (plot F) for the outcome phenotype, each of the ninepairwise combinations of pQTL and outcome association statistics (represented as lines with different colors in the middle of this figure) will be tested using colocalization. In this case, the combination of plot B and plot E shows evidence of colocalization but the remaining eightdo not.