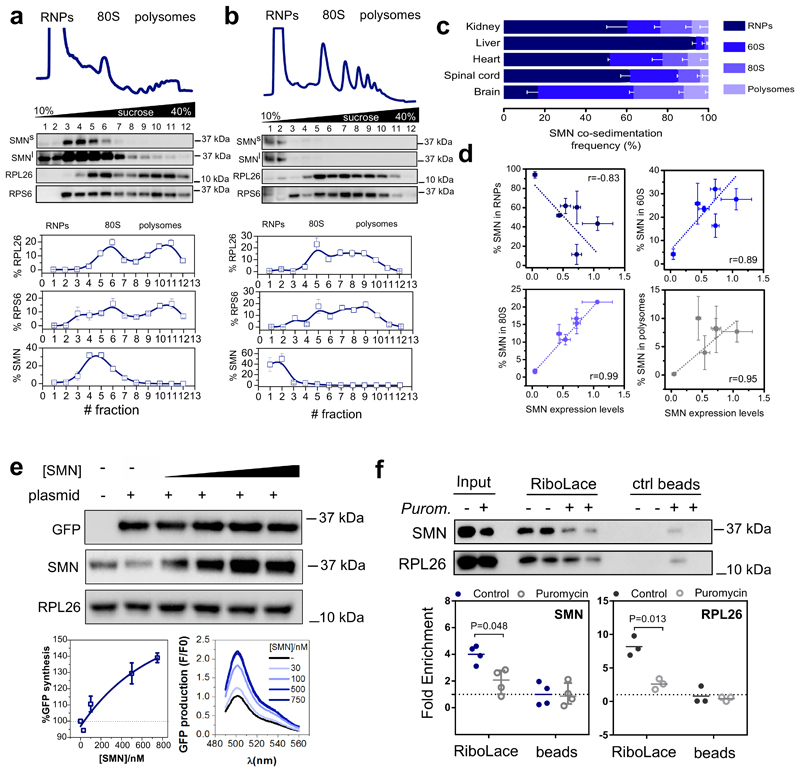
Fig. 2. SMN interacts with the translation machinery in a concentration dependent manner across different tissues and is associated to actively translating ribosomes positively regulating translation.
Co-sedimentation profiles of SMN in a, brain and b, liver. The relative distributions of SMN, and markers of the small (RPS6) and large (RPL26) subunits of the ribosome were used as controls for sedimentation (upper panels). The relative distribution of each protein along the profile is shown as the average ± SEM of n=3 biologically independent experiments. Immunoblots were acquired at short (SMNs) and long (SMNl) exposure time. c, Summary of SMN co-sedimentation with RNPs, 60S, 80S and polysomes in different tissues. The percentages are shown as the average ± SEM of n=3 biologically independent experiments and were obtained using co-sedimentation profiles shown in panels (a, b and Extended data Fig.2b-d). d, Relationship between the relative expression level of SMN in different tissues from (34) and the relative distribution of SMN in RNPs, 60S, 80S and polysomes obtained from (c). Data are presented as the average ± SEM of n=3 biologically independent experiments. e, In vitro translation of reporter GFP in the presence of different concentrations of recombinant SMN. As a negative control a reaction in the absence of the GFP reporter was run in parallel. RPL26 was used as a loading control. Left lower panel, semi-quantitative analysis of GFP level in the presence of different concentrations of recombinant SMN. Plotted are the averages ± SEM from n=3 independent experiments. Right lower panel, the production of GFP was monitored by measuring the appearance of fluorescence in independent assays. f, Western blot analysis of SMN association to active ribosomes using RiboLace (36) in human cells (upper panels) before and after treatment with the translation inhibitor puromycin (100 μM, 1h). RPL26 is used as a marker of ribosomes. The enrichment of SMN and RPL26 with respect to the not-functionalized beads is shown. Plotted are the average ± SEM for n=4 (SMN) and n=3 (RPL26) biologically independent experiments. Significant changes were assessed using a two-sided t-test. Statistical source data and unprocessed blots are provided in Source data Fig. 2.