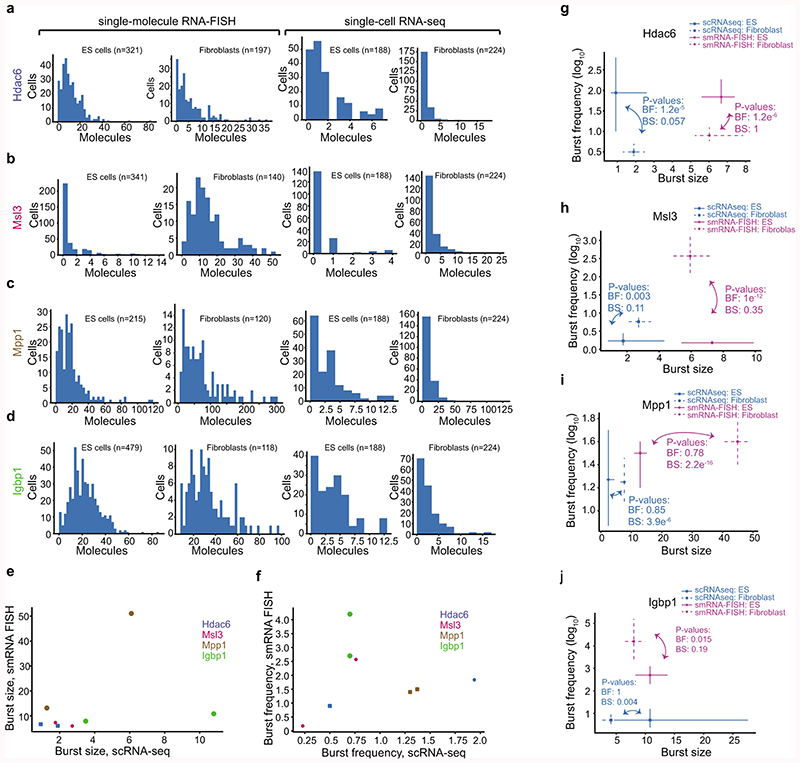
Extended Data Figure 7. Expression distributions and inferred kinetics from single-molecular RNA-FISH and scRNA-seq.
(a-d) Histograms of the expression distributions of genes measured by smFISH and scRNA-seq for genes: Hdac6 (a), Msl3 (b), Mpp1 (c) and Igbp1 (d). Left panel is sm RNA FISH and right panel is scRNA-seq. The number of cells quantified for each gene, cell type and method is presented above each figure item. (e-f) Scatter plots of burst size (e) and frequency (f) inferred based on data from scRNA-seq and smFISH. Data from both fibroblasts and ES cells are shown. Although the number of data points are few and the data do not allow for a systematic comparison between methods, we observed a few trends. There was a good agreement for both burst size and frequency except for the gene Igbp1 that is an outlier in both scatterplots. Igbp1 has increased burst size and lower burst frequency in scRNA-seq compared to smRNA FISH. Excluding Igbp1, we do see a fairly linear correspondence between methods over the remaining 6 data points (3 genes and two cell types). (g-j) Point estimates and confidence intervals shown for each gene, cell type and method based on the profile likelihood method. Number of cells used for the inference is shown in the corresponding histogram in (a-d). P-values for cell-type comparison in burst kinetics is shown per method based on the profile likelihood test.