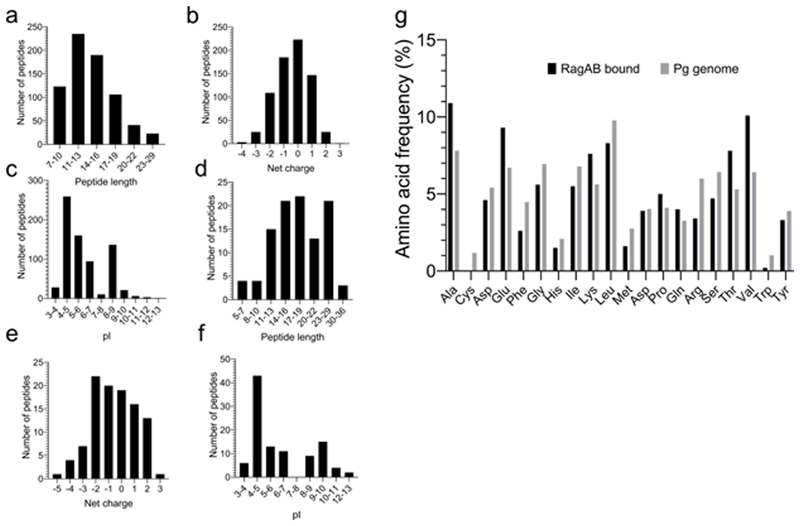
Extended Data Fig. 2. RagAB binds a wide range of oligopeptides.
a-c, LC-MS/MS analysis of peptides bound to RagAB W83 KRAB, showing length distribution (a), total charge (b) and pI (c). d-f, Analysis of peptides bound to RagAB W83 wild-type, showing length distribution (d), total charge (e) and pI (f). For charge calculations, the pH was assumed to be 7.0 and contributions of any His residues were ignored. g, Amino acid frequency of RagAB-bound peptides (KRAB and wild-type combined; black) vs. the amino acid composition in the P. gingivalis proteome (gray), showing a substantial enrichment of Ala, Glu, Lys, Thr and Val. By contrast, aromatics (Phe, Trp) and bulky hydrophobics (Leu) appear to be under-represented. The peptides bound to RagAB from W83 KRAB vary in length from 7 to 29 residues, with a broad maximum of around 13 residues that fits well with the peptide density observed in the structures. Assuming equal abundance of each detected peptide, there is a slight preference for neutral to slightly acidic peptides, and the pI distribution has a bimodal shape, with maxima for acidic and slightly basic peptides. Analysis of the smaller RagAB-bound peptide set from wild-type W83 (d-f) yields a slightly wider size range from 5-36 residues, but overall there are no dramatic differences in the collective length, net charge and pI of the RagAB-bound peptide populations from W83 KRAB and wild-type strains.