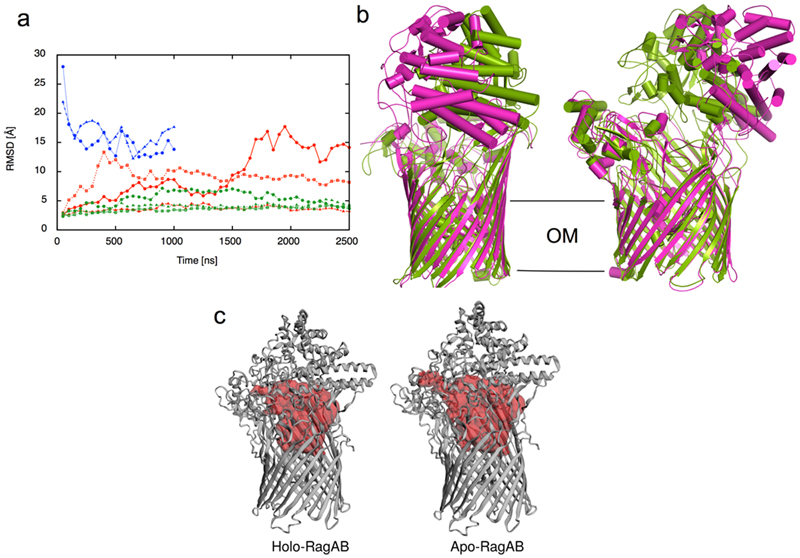
Extended Data Fig. 3. Molecular dynamics simulations of RagAB show lid opening.
a, Cα-rmsd values of RagB lids in apo-RagAB (red) and peptide-bound RagAB (green) with reference to the starting crystal structure in the closed conformation. The Cα-rmsd values of the RagB lids in RagA2B2 are shown in blue with reference to the OO EM state. Each point indicates an average of 50 ns simulation trajectory. b, Comparison of the RagA2B2 open conformation from EM (magenta) with the snapshot of the most open simulation at 2500 ns (green). c, Internal surface of peptide binding cavities in closed holo-RagAB and apo-RagAB, generated with CASTp68. The bound peptide from a RagAB subunit in the crystal structure was removed in silico to generate a closed apo-complex, and three independent MD simulations were performed. For one of the simulations, a clear opening of the RagB lid was observed, reminiscent of recent results for a SusCD transporter and supporting the notion that ligand removal resets the transporter to favour the open state16. None of the peptide-bound complexes shows lid opening on the timescale of the simulations. While this suggests that lid opening is less favourable in the ligand-bound state, it does not contradict our observation of open, ligand-bound complexes via EM. The EM structures allowed us to compare both open states, which showed that the RagB lid in the simulation opens less wide than that in the EM structure, at least during the timescale of the simulation. We also observed a partial closing of both RagB lids during a 1000 ns simulation starting from the OO EM state (a, blue curves). The r.m.s.d. values of both RagB subunits decrease from ~30 Å in the EM structure (t = 0 ns) to ~15 Å, which is similar to the opening observed in one of the apo-RagAB simulations starting from the closed structure. Thus, it appears that the energy minimum for the open state in the simulations is different from that in solution, for reasons that are not clear.