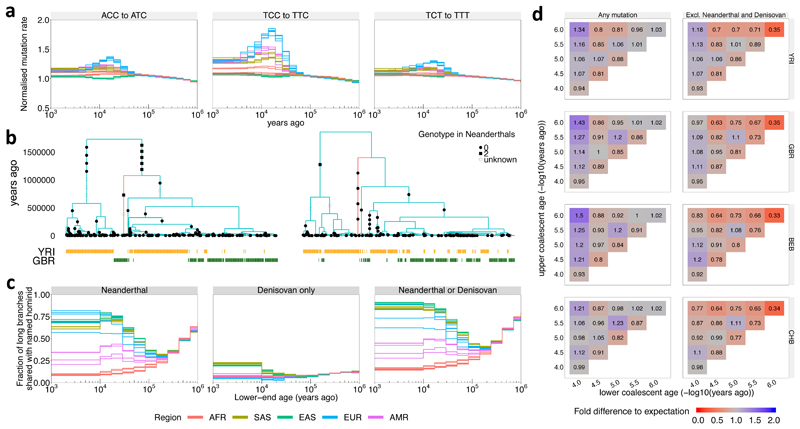
Figure 4. Evolution of human mutation rates and evidence for introgression.
a, Evolution of mutation rates for three triplet mutations ACC to ATC, TCC to TTC, and TCT to TTT (see Supplementary Fig. 7 for all 96 triplet mutations, Methods for normalization). b, Marginal trees for a subregion on chromosome 14 (left) and chromosome 11 (right). The tree on the left contains a long branch with descendants only in GBR (red) consistent with Neanderthal introgression into GBR. The tree on the right contains a long branch with descendants only in YRI (red) consistent with introgression in YRI involving a hominid not closely related to Neanderthals. c, Fraction of branches with an upper-end age >1M YBP that are shared with Neanderthals (left), Denisovans and not Neanderthals (center), or Neanderthals or Denisovans (right) (Methods;). In a and c, colours encode geographic regions (AFR: Africa, EAS: East Asia, EUR: Europe, SAS: South Asia, AMR: Americas). d, Number of mutations binned by age of upper and lower coalescent event, relative to the expected number of mutations when randomising topology while fixing ages of coalescence events (Methods). Right column shows mutations not present in Neanderthal or Denisovan samples.