Figure 5. Natural selection.
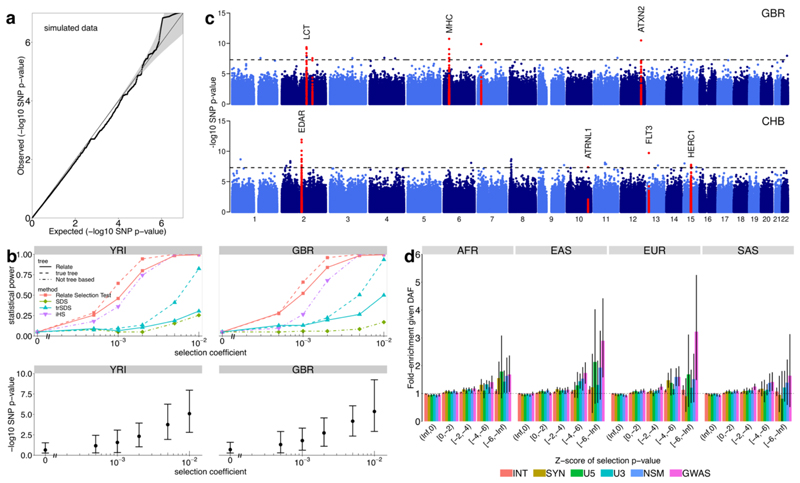
a, QQ-plot of p-values p R for selection evidence of SNPs. We simulated 250Mb for N = 1000 haplotypes using the recombination map of chromosome 1 and a bottleneck population size resembling that of non-African populations. b, Power simulations using N = 1000 haplotypes. We use historical population sizes estimated by Relate for YRI (left) and GBR (right). Top row shows statistical power and bottom row shows p R with the mean indicated by circles and the 5th and 95th percentiles indicated by the error bars. We performed 500 simulations for the neutral case and 200 simulations for s > 0. c, Manhattan plot showing p R of SNPs, for GBR and CHB. We highlight regions containing a SNP with p R < 5 × 10-8 in at least three populations (see Supplementary Table 3 for a full list), as well as the MHC region in GBR. d, Mean enrichment of functional annotation among targets of selection, conditional on allele frequency. Error bars show 95% confidence intervals estimated from 1000 iterations of a block bootstrap resampling (Methods). We group SNPs by mean regional Z-score corresponding to the log p-value for selection evidence, where a smaller Z-score indicates stronger selection evidence. SNPs are binned by partially overlapping functional annotations: intronic mutations (INT), synonymous mutations (SYN), mutations at the 5’ end and 3’ end of a gene (U5, U3), non-synonymous mutations (NSM), and GWAS hits (GWAS).