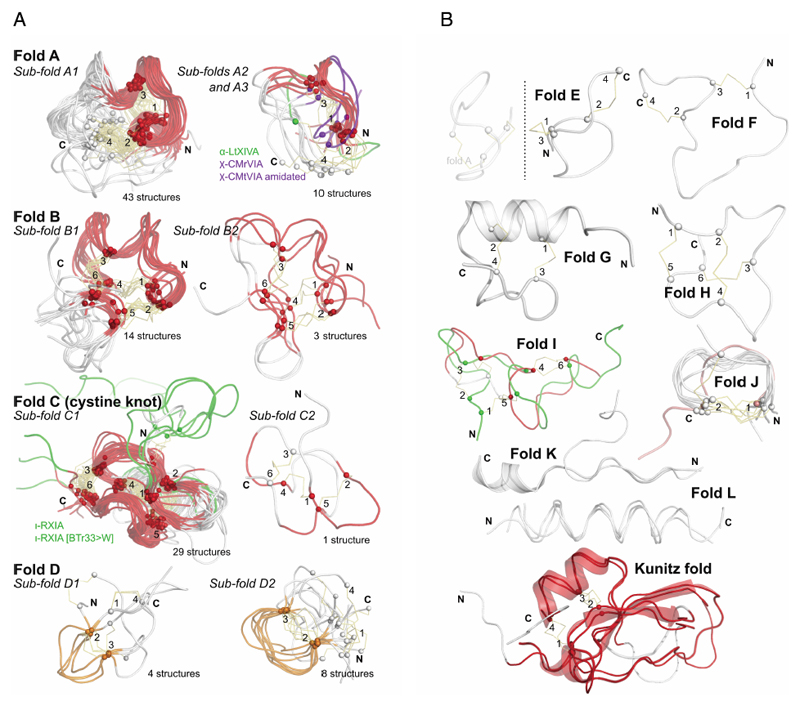
Figure 4.
a: Most commonly discovered or studied conopeptide folds. All available three-dimensional structures in ConoServer corresponding to the four folds A to D were overlaid. The peptide backbone of each conopeptide is shown using a ribbon representation. The alpha carbon of cystine residues or equivalent (i.e. selenocysteines or half-carba-bridge) are represented as spheres, and the cross-links are shown using orange sticks. The most structurally conserved regions are highlighted in red or in orange. Some structures presenting interesting differences to the fold and discussed in the text are colored in green or blue. The half-cystines have been numbered according to their sequential position in the primary sequence, allowing to clearly distinguish the cross-link connectivities. A description of all the structures is provided in Table 4. This figure was partly drawn using PyMol.1
b: Conopeptide folds with only a few representatives. All available three-dimensional structures in ConoServer corresponding to the four folds E to L and Kunitz are overlaid. The peptide backbone structure of each conopeptide is shown using a ribbon representation, and also using a cartoon representation for fold G, K and Kunitz. The alpha carbons of the cystine residues are represented as spheres, and the cross-links are shown using orange sticks. The most structurally conserved regions are highlighted in red for the Kunitz fold. The half-cystines have been numbered according to their sequential position in the primary sequence to clearly distinguish the cross-link connectivities. A description of all the structures is provided in Table 4. The figure was partly drawn using PyMol.1