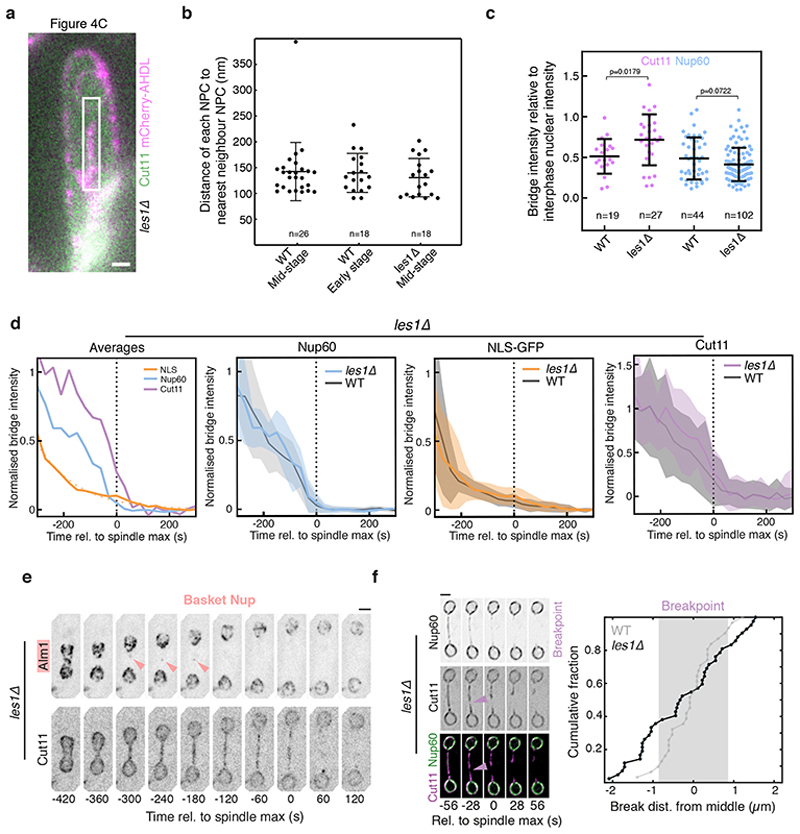
Extended Data Figure 7. Nucleoporin localisation and dynamics in les1Δ cells.
a. Fluorescence image of resin section through a les1Δ cell expressing Cut11-GFP (green) and a synthetic mCherry-AHDL construct (magenta), corresponding to the cell that was imaged by electron tomography shown in Figure 4c. Representative of 2 technical repeats. Scale bar = 1 μm. b. Inter-pore distances for tomograms shown in Figures S3b (WT Mid-stage), 2b (WT Early stage) and tomogram not shown (les1Δ Mid-stage). See Methods for details on measurement. n=26 (WT mid-stage), 18 (WT early stage) and 18 (les1Δ mid-stage) individual nuclear pores in each dataset. The central lines represent the mean of the population, with the error bars representing standard deviation. c. Bridge intensity at bridge formation as an indirect readout of nucleoporin copy number in individual cells, for wild-type (WT) and les1Δ cells. n=19 (WT Cut11), 27 (les1Δ Cut11), 44 (WT Nup60) and 102 (les1Δ Nup60) individual cells pooled from a minimum of 2 biological replicates. Bars overlaid on top of individual data points represent the mean (central line) and standard deviation (error bars). p-values derive from a two-tailed unpaired t-test. See Methods for details. d. Decay curves for NLS-GFP (orange, from 28 cells at t=-300 to 39 cells at t=0), Nup60 (blue, from 10 cells at t=-300 to 15 cells at t=0) and Cut11 (magenta, from 10 cells at t=-300 to 15 cells at t=0) in a les1Δ background, drawn from a minimum of 2 biological replicates. Each trace was normalized by division by maximum bridge signal for that cell prior to averaging. The plot on the left represents the population averages for each marker in a les1Δ background, and the dotted orange line indicates exponential fit to the NLS-GFP average. The 3 subsequent plots show the averages (darker line) and standard deviation (shaded area) for each marker (NLS-GFP, orange; Nup60, blue; Cut11, magenta) in a les1Δ background, overlaid on the wild-type equivalents in gray. e. les1Δ cells expressing Alm1-mNeonGreen and Cut11-mCherry. Arrow indicates a small cluster of Alm1 that enters the bridge but then disappears. Representative of >20 cells from 2 biological repeats. Scale bar = 2 μm. f. SRRF reconstructions of confocal slices at 28 second intervals of les1Δ cells expressing Nup60-mNeonGreen and Cut11-mCherry tagged at the endogenous loci, aligned relative to spindle max (t=0). Representative of >10 cells across 2 technical replicates. Magenta arrow indicates breakpoint. Scale bar = 2 μm. On the right, the cumulative distribution (42 cells from 2 strains across 3 biological replicates) of breakpoint locations relative to the midzone in les1Δ cells. The shaded gray area represents the mean +/- standard deviation of breakpoint locations in wildtype cells (31 cells from 2 biological replicates), with the cumulative distribution as a gray line.