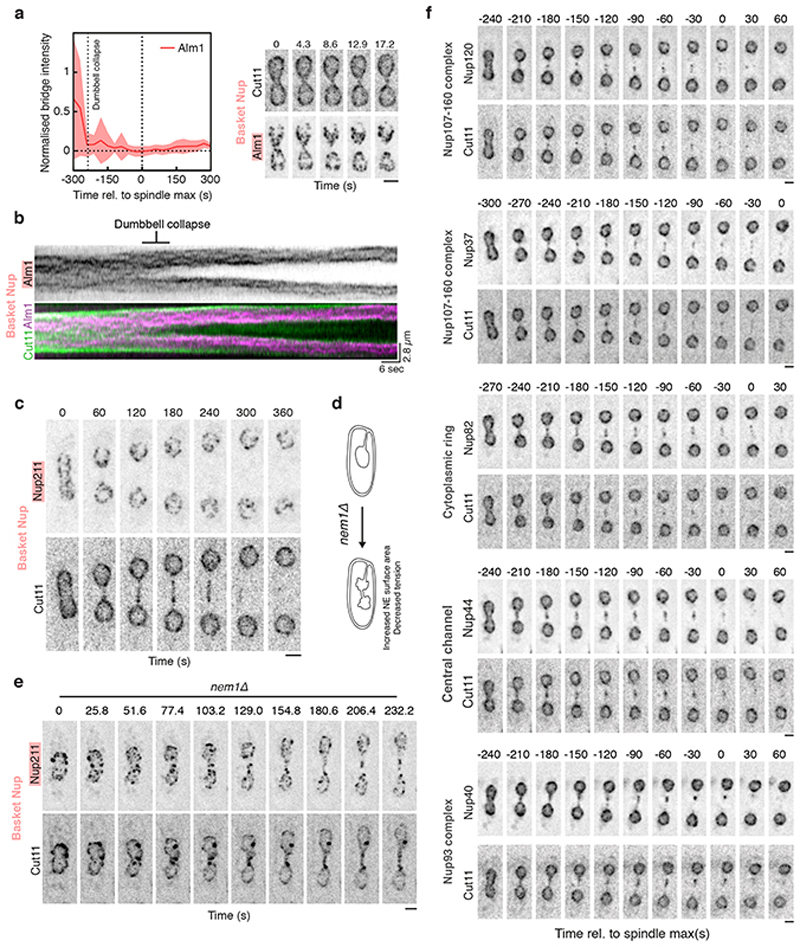
Extended Data Figure 4. Dynamics of individual nucleoporins.
a. Averaged normalised intensity traces for Alm1-mNeonGreen (between 9 cells at t=-300 to 26 cells at t=0) aligned by spindle max (t=0). The central line represents the mean of the population; the shaded area depicts the standard deviation. Images on right are confocal maximum intensity projections of dividing cells expressing Alm1-mNeonGreen and Cut11-mCherry, representative of >20 cells across 3 biological repeats. Scale bar = 2 μm. b. Kymograph of intensities averaged across a single confocal plane of a dividing cell expressing Cut11-mCherry and Alm1-mNeonGreen and imaged at 3 frames per second followed by denoising (see Methods). Representative of >10 cells across 2 technical repeats. c. Confocal maximum intensity projections of dividing cells expressing Nup211-mNeonGreen and Cut11-mCherry. Representative of >30 cells across 3 biological repeats. Scale bar = 2 μm. d. Schematic indicating effect of deleting Nem1 on NE surface area and tension. e. Confocal maximum intensity projections of nem1Δ cells expressing Nup211-mNeonGreen and Cut11-mCherry. Representative of >30 cells across 2 biological repeats. Scale bar = 2 μm. Note that the basket Nup is able to enter the nuclear bridge in this condition. f. Confocal maximum intensity projections of dividing cells expressing various NPC subcomplex components tagged with mNeonGreen at their C termini, co-expressed in each case with Cut11-mCherry. Each set of panels is representative of >20 cells across 2 biological repeats. Scale bars = 2 μm.