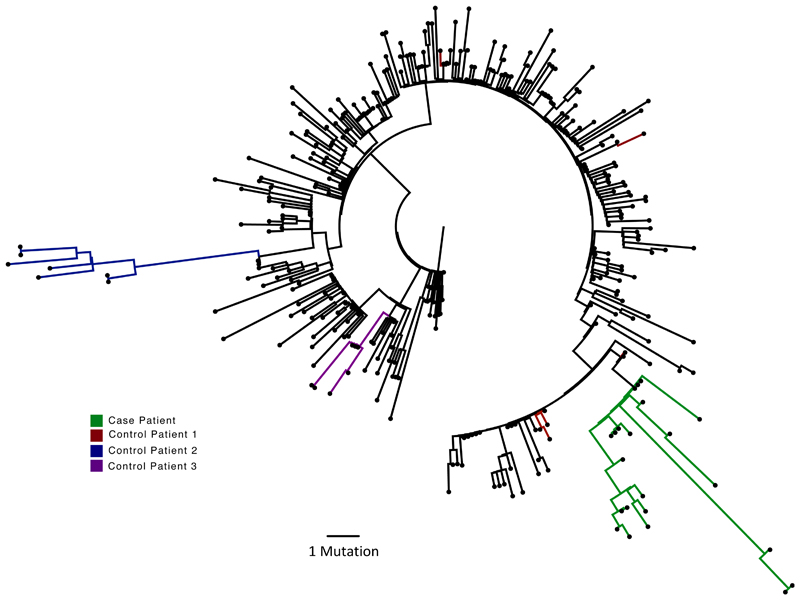
Figure 1.
Analysis of 23 Patient derived whole SARS-CoV-2 genome sequences in context of local sequences and other cases of chronic SARS-CoV-2 shedding. Circularised maximum-likelihood phylogenetic tree rooted on the Wuhan-Hu-1 reference sequence, showing a subset of 250 local SARS-CoV-2 genomes from GISAID. This diagram highlights significant diversity of the case patient (green) compared to three other local patients with prolonged shedding (blue, red and purple sequences). All “United Kingdom / English” SARS-CoV-2 genomes were downloaded from the GISAID database and a random subset of 250 selected as background.