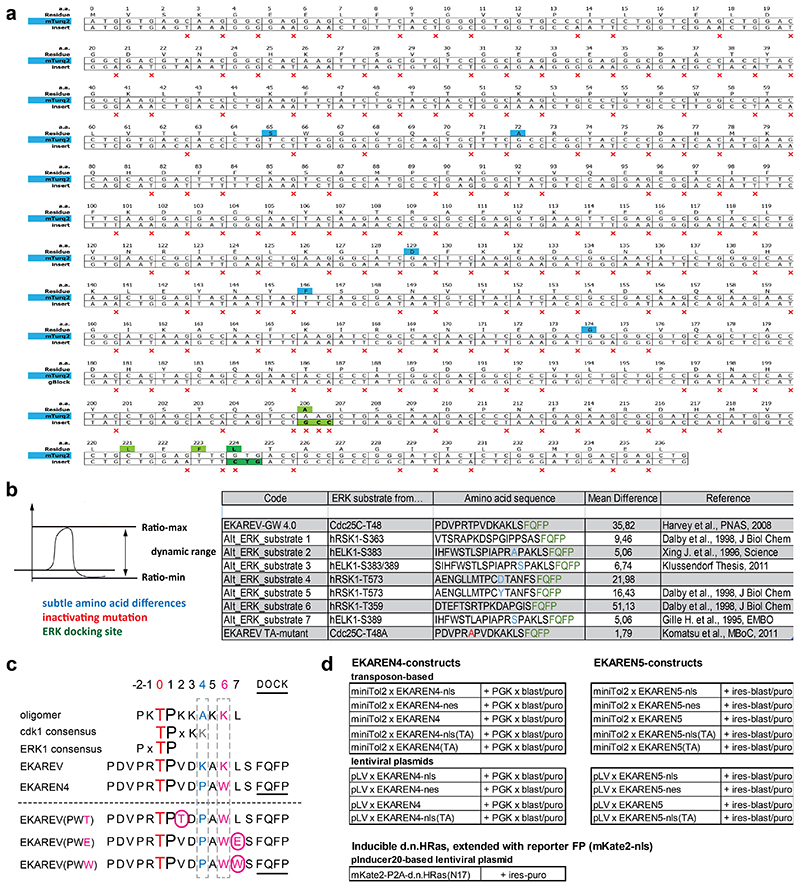
Extended Data Fig. 4. Biosensor sequences.
a, Silent mutations introduced into Turquoise2 (insert) to minimize sequence homology with the YPet fluorophore in the same construct. Red ‘x’ marks silent mutations. Blue, residues discriminating Turquoise2 from parental eCFP (Goedhart et al., 2012). Green, residues rendering Turquoise2 prone to dimerize with YPet, in analogy to EKAREV design (Komatsu et al., 201128). Dark green, the V224L mutation was added to further enhance dimerization and, hence, FRET efficiency in ON-state (Vinkenborg et al., 200732).
b, Alternative ERK substrate sequences were derived from ERK targets RSK1 (human) and ELK1 (human) using the Kinexus website and compared to parental EKAREV-GW-4.0 (GW = GateWay) with CDC25C substrate sequence.
c, Overview of generated and tested point mutant variants of EKAREV. Red, central Threonine, target of ERK phosphorylation. Blue, the Lysine at position +4 mimics the general CDK1-consensus site, mutated to Proline (K->P). Purple, the Lysine at position +6 mimics the CDK1-consensus site and mutated to bulky Trp (K->W) to create steric hindrance with cyclinB. Underlined, ERK docking domain FQFP. Purple boxed characters, rational attempts to further eliminate CDK1-sensitivity with third amino acid replacements. V422T was aimed at favoring ERK over CDK1 consensus site; L427W was aimed at further augmenting sterical hinderance of cyclinB interaction; L427E was aimed at impeding cyclinB interaction through electrostatic repulsion. The Asp (D) at position+3 was left unchanged for its reported importance for the Pin1 affinity (Komatsu et al., 201128).
d, Summary of available EKAREN4 and EKAREN5 plasmid constructs (including variable targeting motifs and Thr-Ala control versions), as well as adapted version of pInducer20 (Meerbrey et al., 201145) to initiate expression of HRASN17 and P2A-coupled reporter fluorophore mKate2-NLS.