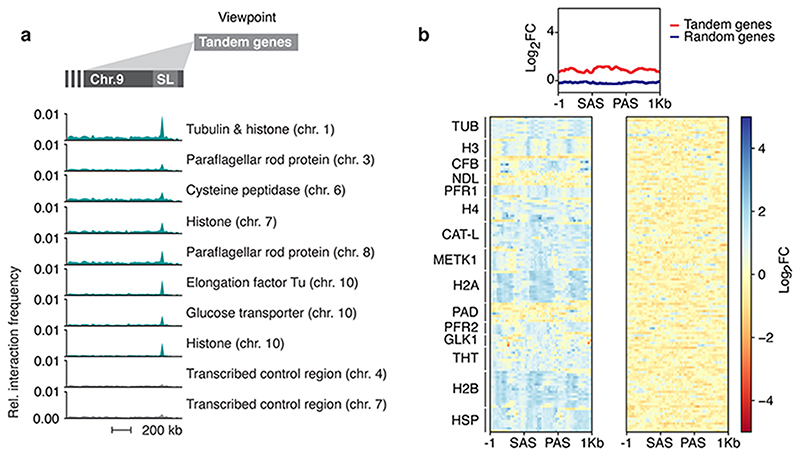
Extended Data Fig. 9. Tandem arrays interact with the Spliced Leader (SL) locus.
a, Hi-C (virtual 4C) analysis, viewpoint: different tandem gene arrays and control sites. Relative interaction frequencies between the different viewpoints and the SL-RNA locus are plotted. Bin size 20 kb. As viewpoint control regions, two actively transcribed regions from the diploid cores of chr. 4 and chr 7 were arbitrarily chosen. The coordinates of all viewpoints used for virtual 4C analyses are listed in Supplementary Information sheet 3. b, VEX1myc chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) analysis. Top panel, metagene plot for tandem genes compared to randomly selected genes with no paralogues. Lower left, heat map of tandem genes. Lower right, randomly selected genes with no paralogues.