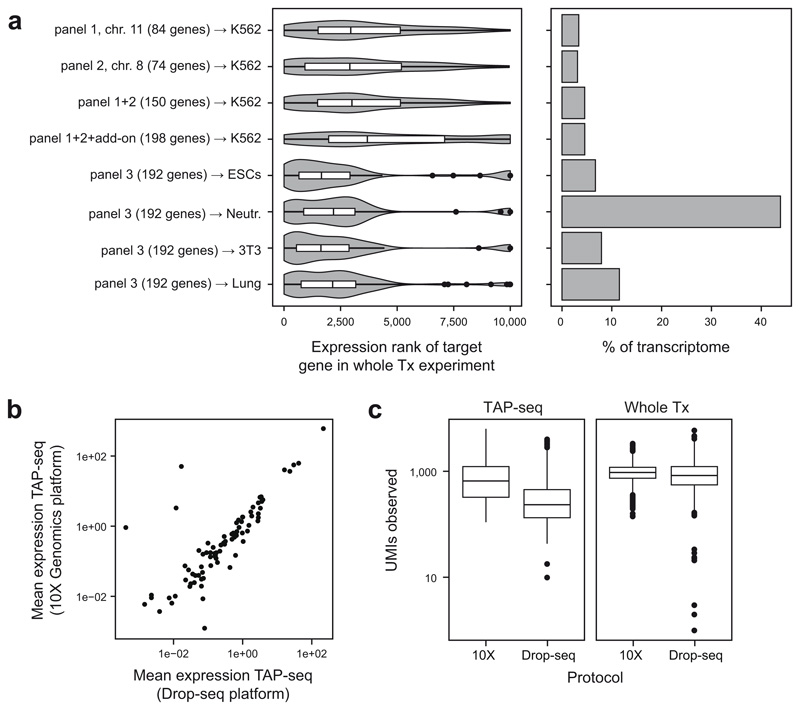
Extended Data Fig. 2. Choice of target genes and single cell capture platform.
a. Left panel: Expression ranks of genes used for the three test panels. An x-axis value of 1 refers to highest expression. See y-axis labels for number of target genes, and refer to Methods section on ‘data visualization’ for a description of violin plot elements. Right panel: Fraction of the transcriptome covered by these panels, computed from a whole transcriptome reference data set from the same cell type (y-axis labels). For panels 1+2, K562 cells were used, panel 3 was applied to mouse embryonic stem cells (ESCs), mouse 3T3 cells, mouse neutrophils (Neutr.), and mouse lung mesenchymal cells (Lung). b. Mean gene expression levels for 10X Genomics and Drop-seq based TAP-seq. Panel 1 was used in both cases. n=84 genes are shown. c. Number of UMIs observed per cell for both TAP-seq and whole transcriptome readout (Whole Tx), using 10X Genomics or Drop-seq for RNA capture and reverse transcription. Experiments were downsampled to an average of 1,000 reads per cell.