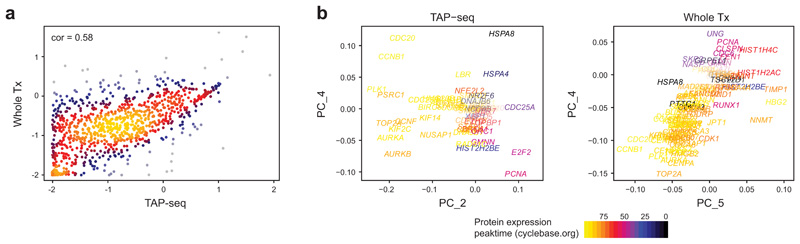
Extended Data Fig. 3. Analysis of the L1000 panel.
a. Spearman correlation in mean gene expression levels between TAP-seq and whole transcriptome readout (Whole Tx) for a panel targeting the L1000 gene set34. n=963 genes were covered in TAP-seq and are included in the plot. b. Principal component analysis of the TAP-seq dataset and the whole transcriptome dataset. Principal component loadings of all genes annotated in cyclebase v352 are shown, with the peak-time of expression color-coded. In the whole transcriptome dataset, PC1-3 were not significantly associated with GO-terms (not shown). n=8734 cells (TAP-seq) and 8282 cells (Whole Tx).