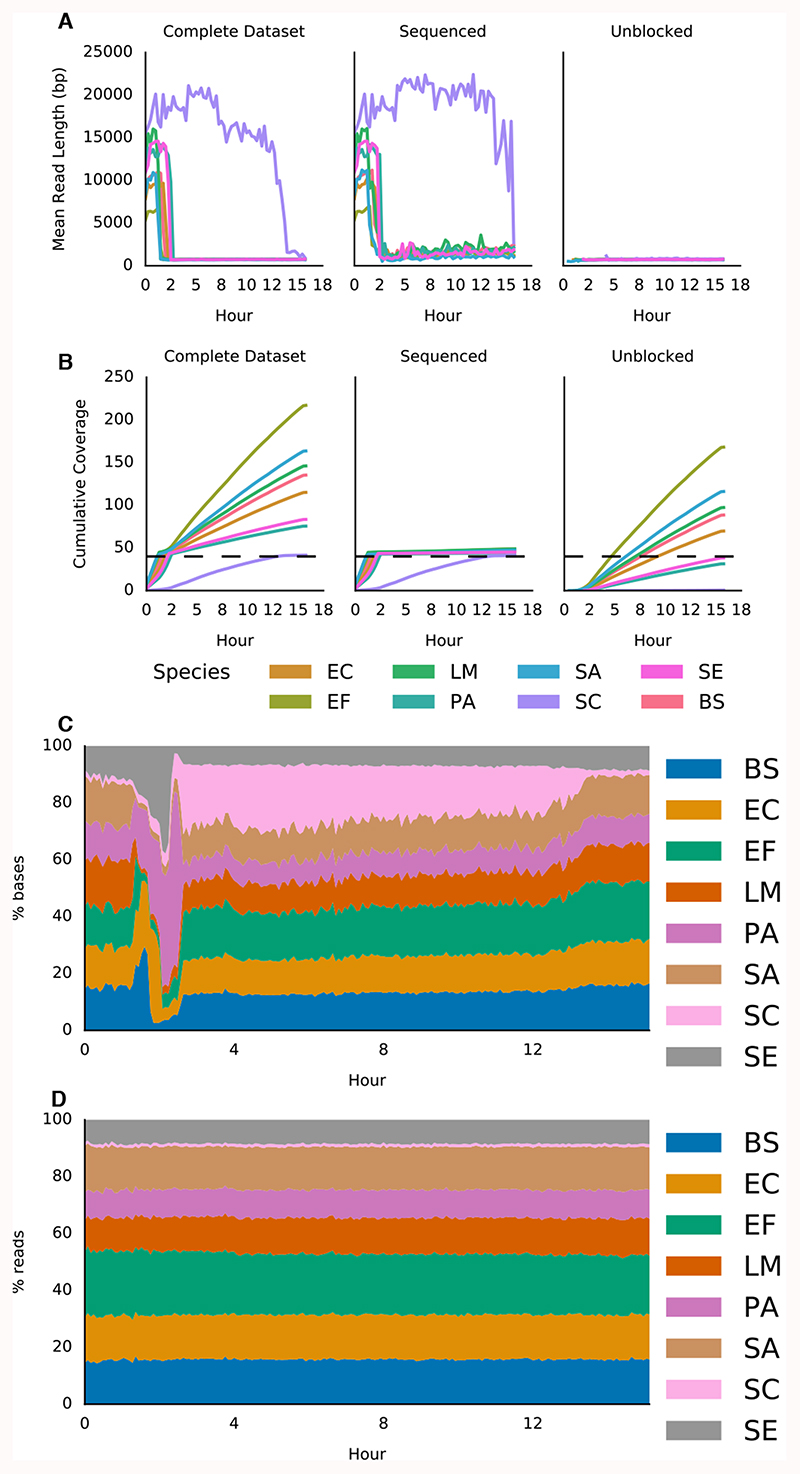
Figure 2. Adaptive sequencing enriching for the least abundant genome and ensuring uniform 40x coverage.
A) Mean read lengths for reads sequenced from the ZymoBIOMICS mock metagenomic community mapped against the provided references (ZymoBIOMICS, USA). Read lengths are reported for the whole run, the deliberately sequenced reads and those which were actively unblocked. B) Shows cumulative coverage of each ZymoBIOMICS genome during the sequencing run. The total coverage still accumulated as unblocked reads, though short, still map. Sequencing was automatically terminated once each sample reached 40x. C) Stacked area graph illustrating how the proportion of bases mapping to each species changes over time. D) In contrast, the proportion of reads mapping to each species over time doesn’t change significantly. Species and composition are: bs - Bacillus subtilis (14%), ef - Enterococcus faecalis (14%), ec - Escherichia coli (14%), lm - Listeria monocytogenes (14%), pa - Pseudomonas aeruginosa (14%), sc - Saccharomyces cerevisiae (2%), se - Salmonella enterica (14%), sa - Staphylococcus aureus (14%).