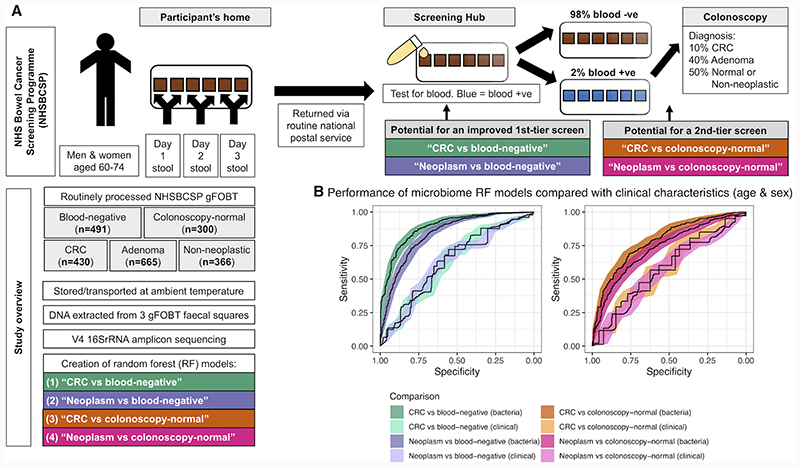
Figure 1. Microbiome taxonomic profiling demonstrates potential to improve CRC screening accuracy.
(A) Overview of the NHS Bowel Cancer Screening Programme (NHSBCSP) and the design of this study. Briefly, we used 16S amplicon-based microbiome profiling from routinely collected gFOBT specimens to supplement first-tier (CRC/neoplasm vs. blood-negative) or second-tier (CRC/neoplasm vs. colonoscopy-normal) opportunities for early cancer screening. (B) Microbiome profiles improve CRC or neoplasm classification versus blood-negative gFOBT samples (first-tier screening application) or blood-positive colonoscopy-normal samples (second-tier screening application) relative to purely clinical characteristics (age and sex). Classification used random forest (RF) models and shows the performance of the ‘total’ RF models bootstrapped from the total datasets. Shading represents the 95% CI. Clinical = RF models based on age & sex. Bacteria = RF models based on relative abundances of genera. Neoplasm = a group comprising an approximately equal ratio of CRC, low-risk adenoma, intermediate-risk adenoma and high-risk adenoma samples.