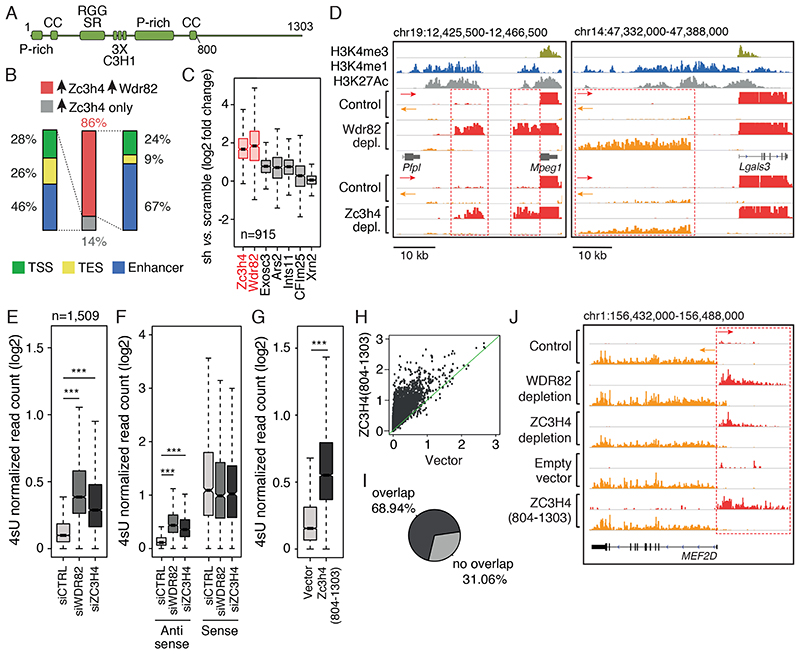
Figure 1. Effects of ZC3H4 depletion on extragenic transcription.
A) Schematic representation of the ZC3H4 protein with annotated domains. P-rich: proline-rich region; CC: coiled-coil; RGG: Arginine-Glycine-rich domain; SR: Arginine-Serine dipeptide-rich motif; C3H1: CCCH type Zn fingers.
B) Overlap between extragenic transcripts upregulated upon ZC3H4 depletion (n=915) and those upregulated upon WDR82 depletion (n=3) in mouse macrophages. Genomic annotation of transcripts concordantly upregulated upon depletion of ZC3H4 and WDR82 or transcripts upregulated only upon depletion of ZC3H4. TSS = Transcription Start Site; TES = Transcription End Site. Data from n=3 independent experiments are shown.
C) Effects of the depletion ZC3H4 and other termination factors on transcription of extragenic regions suppressed by WDR82 in mouse macrophages. Data from n=3 independent experiments are shown.
D) Representative genomic regions showing the effects of WDR82 and ZC3H4 depletion on extragenic transcription in macrophages. Red and orange tracks correspond to plus and minus strand RNAs, respectively.
E) Upregulation of extragenic transcription in HeLa cells depleted of WDR82 or ZC3H4 by siRNA transfection. Read counts at n=1,513 extragenic regions upregulated in HeLa cells depleted of WDR82 are reported. Data from n=3 independent experiments are shown. Statistical significance was assessed using the two-tailed Wilcoxon signed rank test (p-value < 2.2e-16 in both comparisons). *** = p-value <0.01.
F) Promoter-antisense RNAs (n=429) upregulated in 4sU-seq data sets upon WDR82 or ZC3H4 depletion in HeLa cells. Transcription of the paired sense (coding) RNAs is shown. Data from n=3 independent experiments are shown. Statistical significance was assessed using the two-tailed Wilcoxon signed rank test for the comparison between siWDR82 vs. siCTRL (p-value=1.9e-210) and siZC3H4 vs siCTRL (p-value=1.1e-203). *** = p-value <0.01.
G) Effects of ZC3H4(804-1303) overexpression on transcription of the n=1,509 extragenic regions upregulated in WDR82-depleted HeLa cells. Data from n=3 independent experiments are shown. Statistical significance was assessed using the two-tailed Wilcoxon signed rank test (p-value < 2.2e-16).
H) Same data as in panel G were represented as scatter plot.
I) Overlap of the transcripts upregulated upon over-expression of ZC3H4(804-1303) and the transcripts upregulated in HeLa cells depleted of WDR82 or ZC3H4.
J) A representative genomic region showing MEF2D sense and promoter-antisense transcription in HeLa cells depleted of WDR82 or ZC3H4 or over-expressing ZC3H4(804-1303).
For panels C, E and F, the median value for each fold change is shown with a horizontal black line. Boxes show values between the first and the third quartile. The lower and upper whisker show the smallest and the highest value, respectively. Outliers are not shown. The notches correspond to ~95% confidence interval for the median.