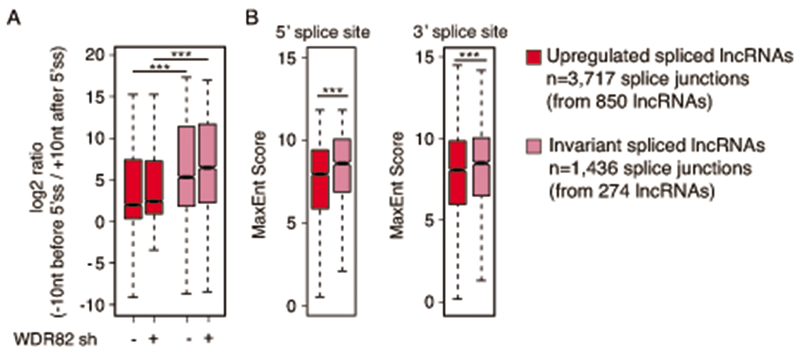
Extended Data Fig. 9. Characterization of splicing efficiency and splice site quality of extragenic transcripts not affected by WDR82 depletion.
A) log2-transformed ratio of polyA RNA-seq reads upstream and downstream of the 5’ splice sites of WDR82-suppressed and WDR82-insensitive lncRNAs in HeLa cells. Statistical significance was assessed using the two-tailed Wilcoxon rank sum test (p-value= 1.8e-175 for the controls and p-value=1.3e-199 in Wdr82-depleted cells). ***p-value < 0.01. Data are from n=3 independent experiments.
B) Analysis of 5’ (left) and 3’ (right) splice site strength at WDR82-suppressed and WDR82-insensitive lncRNAs in HeLa cells. MaxEnt scores for both donor and acceptor splice sites were measured. Statistical significance was assessed using the two-tailed Wilcoxon rank sum test in correspondence of both the 5’ (p-value= 1.3e-20) and the 3’ (p-value= 3e-04) splice sites. *** p-value < 0.01. Data are from n=3 independent experiments.
Boxes show values between the first and the third quartile. The lower and upper whisker show the smallest and the highest value, respectively. Outliers are not shown. The notches correspond to ~95% confidence interval for the median.