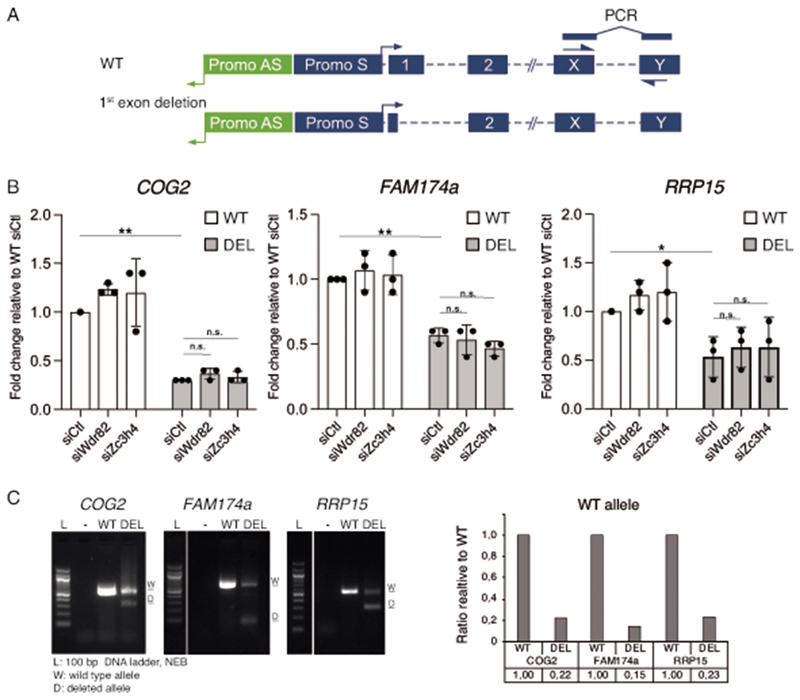
Extended Data Fig. 10. First exon deletions in protein coding genes.
A) Schematic representation of the deletion of the first exons of protein coding genes. sgRNAs were designed to remove a genomic sequence that included the first exon from 30-50nt downstream of the TSS to the intronic sequences just downstream of the 5’ splice site.
B) Expression of the indicated gene mRNAs was measured by qRT-PCR in bulk populations of wild type or first exon-deleted HeLa cells after transduction of the indicated siRNAs. Primers used were specific for spliced mRNAs and were designed on downstream exons (Methods). The plot shows the mean s.d. of n=3 independent experiments. * P < 0.05; **P < 0.01, by two tailed t-test. The data were normalized on the housekeeping gene NRSN2. P-values for COG2: WT vs. DEL siCtl = 1.75E-07; DEL siCtl vs. siWdr82 = 0.78 (n.s.); DEL siCtl vs. siWdr82 = 0.80 (n.s.). P-values for FAM174a: WT vs. DEL siCtl = 0.0005; DEL siCtl vs. siWdr82 = 0.85 (n.s.); DEL siCtl vs. siWdr82 = 0.15 (n.s.). P-values for RRP15: WT vs. DEL siCtl = 0.013; DEL siCtl vs. siWdr82 = 0.70 (n.s.); DEL siCtl vs. siWdr82 = 0.65 (n.s.)
C) First exon deletion efficiency at the three genes tested was analyzed by genomic PCR. The quantification of the wild type allele gel band in wt cells and cells in which the first exon was deleted using sgRNAs+Cas9 is shown on the right. Uncropped images are available online as source data.